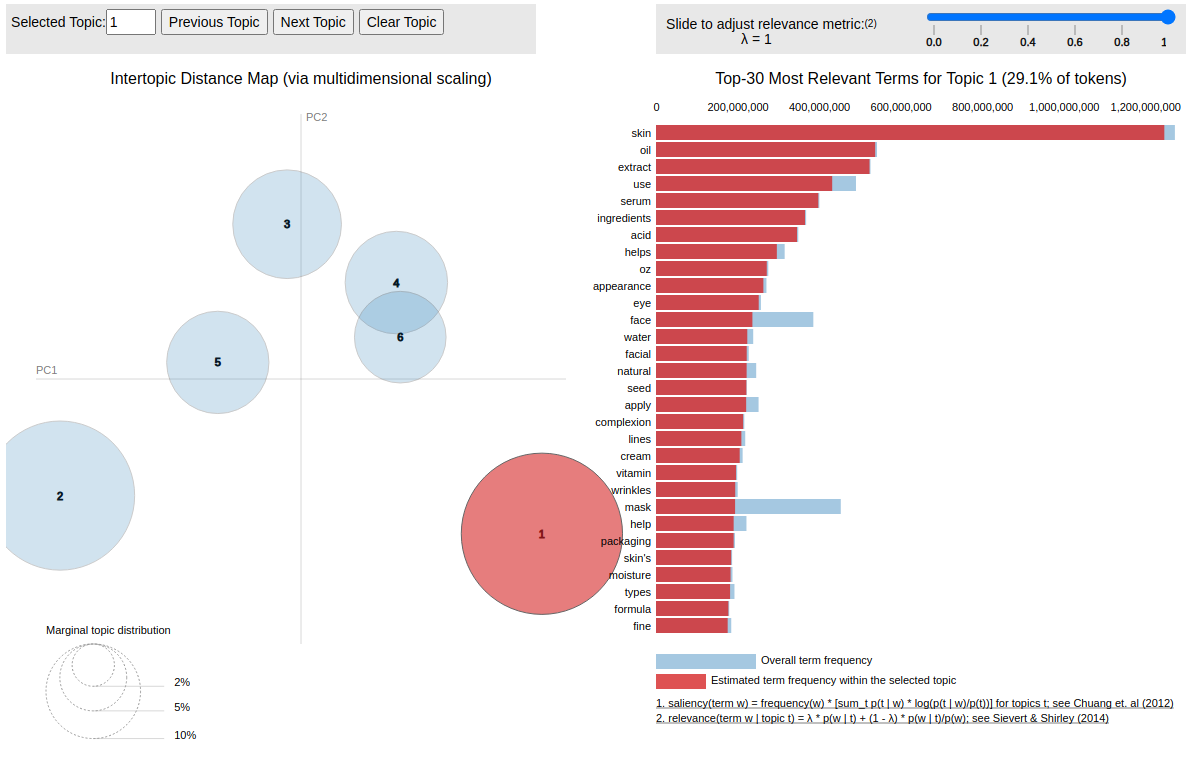
see the detailed report in notebook here
Submitting the job to EMR is easy
RUN: bash emr.sh
Under the hood following things happened:
- Python code including settings will be packaged into local
./distfolder as EMR job artifacts (see more in package.sh) - Local
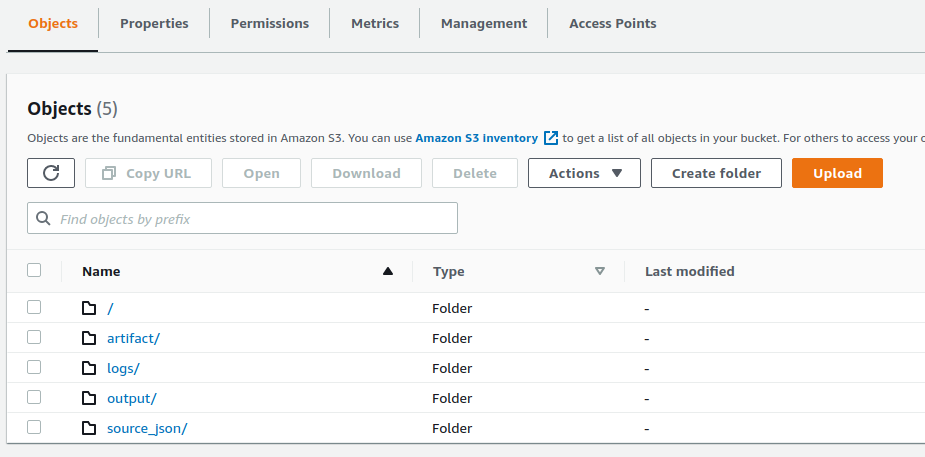
./distthen is uploaded to s3 underartifactprefix. (later referenced by EMR step) - Create new EMR cluster, it read raw data from
source_json, run ETL job - Finally, it saves outputs (such as, transformed data, model, and visualization data files) under
outputprefix in s3.
see s3 folder structure: (logs is EMR log folder)
- Read raw json file from source folder
source_jsonin s3 - Data clean and Feature engineering on product description:
- remove html markups
- remove punctuations and special characters
- remove numbers
- remove common stop words
- then tokenize and vectoring product description corpus
- Fitting feature data via LDA model
- Save transformed data into s3
outputfolder - Save model artifacts
- Save reports for data visualization
- aws accounts
- credentials with emr/s3/airflow policy attached
- pyspark dev environment
- python,tox
Create virtual dev environment via tox
RUN: tox -e dev
ACTIVATE: :{your project root}$ source .tox/dev/bin/activate
OPEN NOTEBOOK: jupyter notebook
Happy coding!
tox -e test