A little script to find enriched functions (pfam/ipr/gene family etc) on chromosomes, eg. fist to make 50-gene windows, and test in each window functional enrichment compared to all genes.
NOTE: the funcitonal genes are not necessarily adjacent for their gene orders.
!!!sorted by chr and coord
Smp_329140 SM_V7_1 88327
Smp_315690 SM_V7_1 103403
Smp_317470 SM_V7_1 256087
Smp_000020 PF07555
Smp_000040 PF13374,PF13424
Smp_000050 PF00520
PF00001 7tm_1
PF00002 7tm_2
SM_V7_1 88881357
SM_V7_2 48130368
...
To get significantly enriched clusters for each block, run
./getClusters_nonSliding.sh [WINDOWSIZE]
- fisher_enriched_nonsliding.txt (significant clusters on each chromosome)
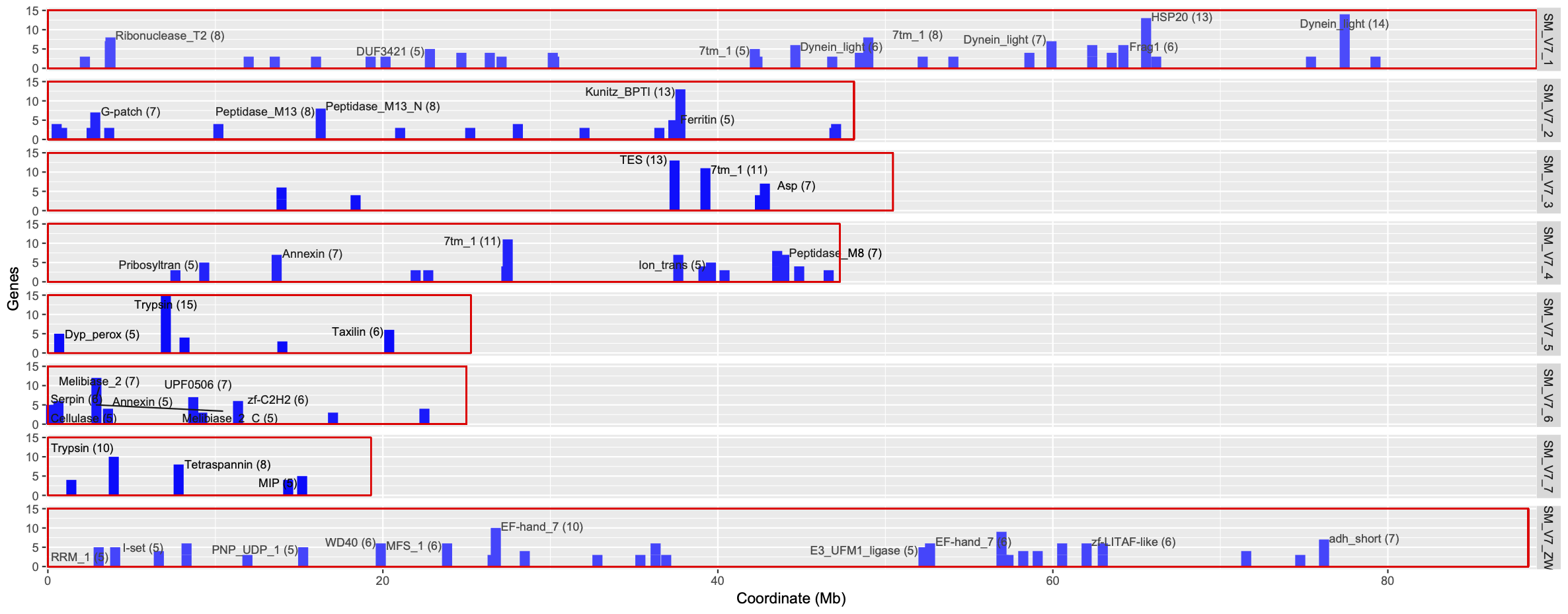
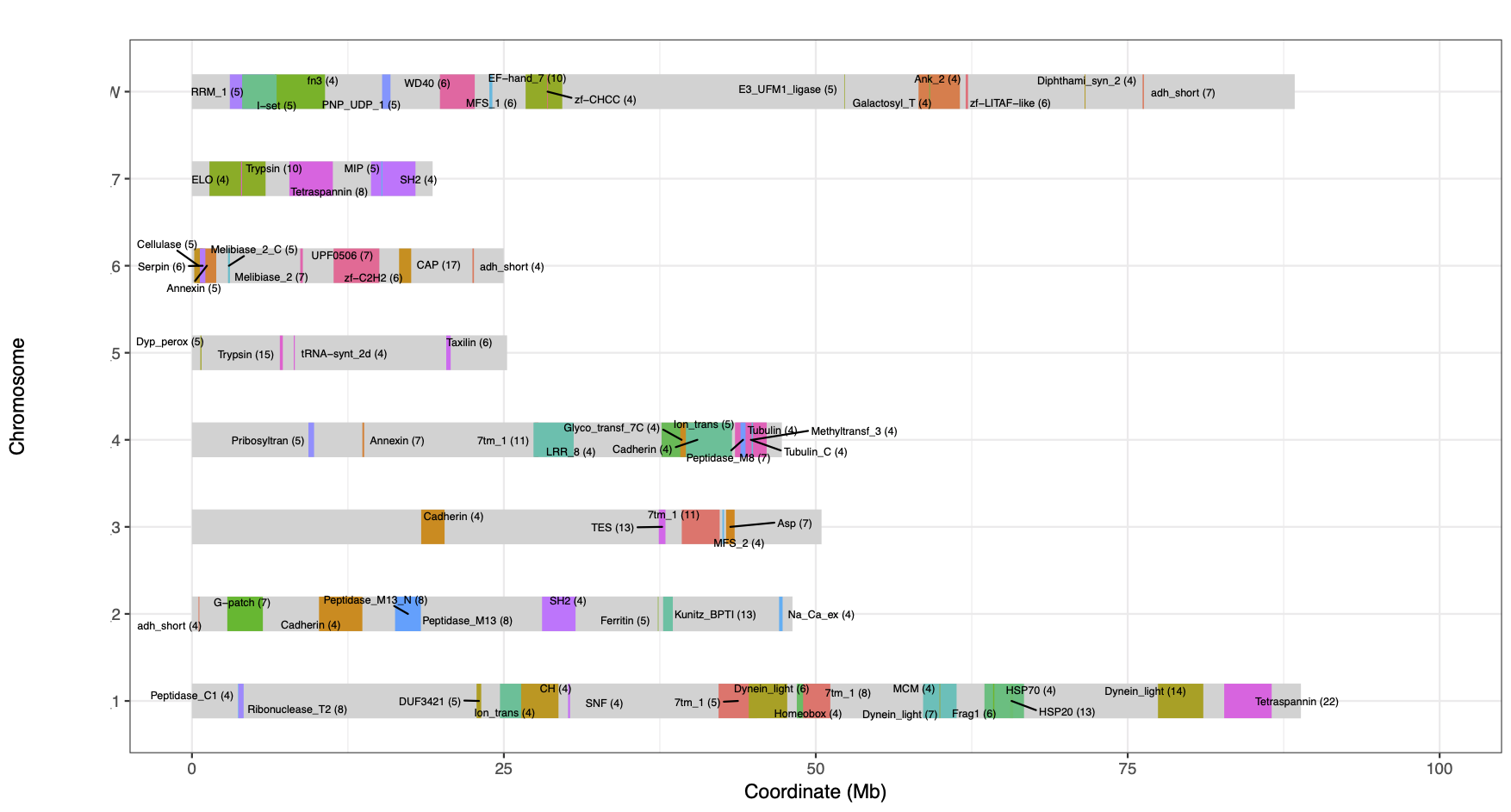
- plot_func-clusters_nonsliding.txt (Significantly enriched functions: FDR<0.05, at least 3 genes)
- plot_func-clusters_nonsliding.txt.pdf (chromosome plots with significant clusters)
Using 50-gene window with stepping 5-genes. Run
./getClusters_Sliding.sh [WINDOWSIZE] [STEPSIZE]
- fisher_enriched_raw.txt (raw test results for all functions and all blocks: overlapping exists)
- fisher_enriched_sliding.txt (significant clusters on each chromosome)
- plot_func-clusters_sliding.txt (Significantly enriched functions: FDR<0.05, at least 3 genes)
- plot_func-clusters_sliding.txt.pdf (chromosome plots with significant clusters)
Note that the current script is not perfect to choose the cluster with most genes. Duplicate clusters may exist and several clusters of the same function may exist but not picked. Suggest to run the non-sliding approach for a reference.
- 3-cluster_geneCounts.R: plot the gene counts in each cluster as a bar
- 4-cluster_geneCoord.R: plot clusters with start coordinates of the first and last genes. Shown as coloured blocks.