AbunRNA
Description
AbunRNA is an R package focuses on the commonly used RNAseq analysis
pipelines including indexing, quantification, and to the downstream
expression analysis. This packages is friendly to users with lack or no
experience in command line usage, but can still access the
high-efficiency sequence mapping software i.e. Salmon. The R-based
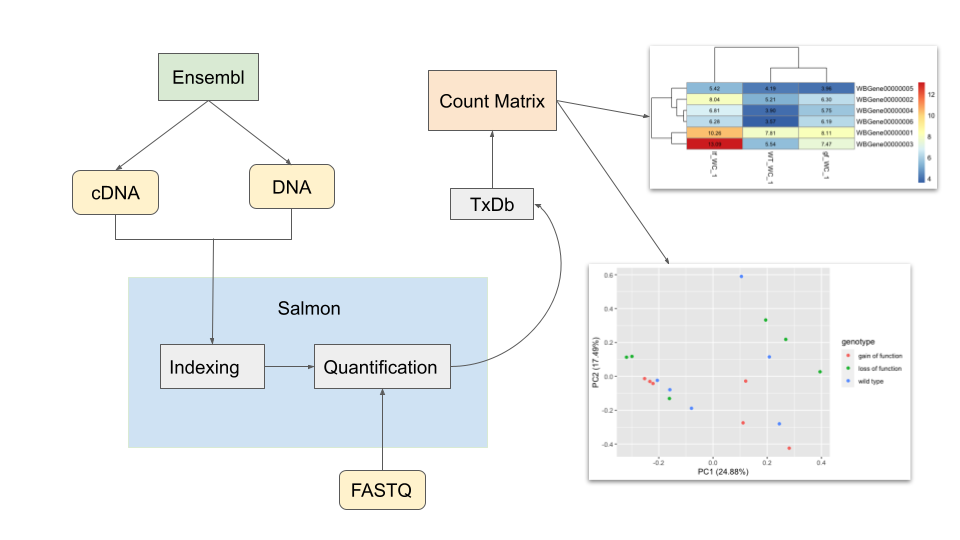
pipeline can access the available cDNA, DNA and transcriptome data from
Ensembl, generate a count matrix indicating the expression abundance of
each genes within each sample, plot a heatmap to visualize the abundance
among wild type and mutants, and finally perform principle component
analysis on the count matrix for more exploratory information. The
AbunRNA package is developed using R version 4.2.1 (2022-06-23),
Platform: x86_64-apple-darwin17.0 (64-bit) and
Running under: macOS Ventura 13.0.
Installation
To install the latest version of the package:
require("devtools")
devtools::install_github("zhangchengyue/AbunRNA", build_vignettes = TRUE)
library("AbunRNA")To run shinyApp:
AbunRNA::runAbunRNA()Sometimes errors about dependencies are showing up, usually that would happen for the following packages not previously downloaded in your RStudio: tximport, biomaRt, GenomicFeatures, AnnotationDbi.
To solve this, run the following code to download these packages first, then re-install AbunRNA.
if (! requireNamespace("BiocManager", quietly = TRUE)) {
install.packages("BiocManager")
}
if (! requireNamespace("tximport", quietly = TRUE)) {
BiocManager::install("tximport")
}
if (! requireNamespace("biomaRt", quietly = TRUE)) {
BiocManager::install("biomaRt")
}
if (! requireNamespace("GenomicFeatures", quietly = TRUE)) {
BiocManager::install("GenomicFeatures")
}
if (! requireNamespace("AnnotationDbi", quietly = TRUE)) {
BiocManager::install("AnnotationDbi")
}
if (! requireNamespace("ggfortify", quietly = TRUE)) {
install.packages("ggfortify")
}Overview
ls("package:AbunRNA")
data(package = "AbunRNA") AbunRNA contains functions to:
- install the popularly used Salmon software through bioconda (See function installSalmon)
- directly obtain reference transcriptome from Ensembl database (See function obtainCDNA, obtainDNA, and obtainGTF)
- run quantification through Salmon (See function quantification)
- generate TxDb object for storing transcript annotations (See function txdbObj)
- generate the count matrix from TxDb and optionally plot the heatmap to visualize abundance (See function generateMatrix)
- performs principle component analysis for the matrix, and plot the graph to further visualize the variation in expression abundance among wild type and mutant samples.(See function plotPCA)
browseVignettes("AbunRNA")
An overview of the package is illustrated below.
Contributions
The author of the package is ChengYue Zhang. The obtainGTF(),
obtainDNA() and obtainCDNA() functions makes use of rvest package
to web scrape from online database. The txdbObj function makes use of
makeTxDbFromGFF() function from GenomicFeatures package to create a
txdb object from GTF file. The generateMatrix() function uses pheatmap
function from pheatmap R package to plot the heatmap. The stats and
ggplot2 R packages are used for principle component analysis in
plotPCA() function. The rstudioapi CRAN package is used to send
commands to terminal for downloading Salmon software in
installSalmon() function, as well as running Salmon commands for
indexing and quantification in quantification() function.
References
Patro, R., Duggal, G., Love, M. et al. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14, 417–419 (2017).https://doi.org/10.1038/nmeth.4197
Ushey K, Allaire J, Wickham H, Ritchie G (2022). rstudioapi: Safely Access the RStudio API. https://CRAN.R-project.org/package=rstudioapi
Ensembl 2022, Nucleic Acids Research, Volume 50, Issue D1,7 January 2022, Pages D988–D995, https://doi.org/10.1093/nar/gkab1049
Durinck S, Spellman P, Birney E, Huber W (2009). “Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt.” Nature Protocols, 4, 1184–1191.
Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W (2005). “BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis.” Bioinformatics, 21, 3439–3440.
Wickham H, François R, Henry L, Müller K (2022). dplyr: A Grammar of Data Manipulation. https://CRAN.R-project.org/package=dplyr
Müller K, Wickham H (2022). tibble: Simple Data Frames. https://CRAN.R-project.org/package=tibble
Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan M, Carey V (2013). “Software for Computing and Annotating Genomic Ranges.” PLoS Computational Biology, 9. doi: 10.1371/journal.pcbi.1003118, http://www.ploscompbiol.org/article/info%3Adoi%2F10.1371%2Fjournal.pcbi.1003118
Wickham H (2022). rvest: Easily Harvest (Scrape) Web Pages. https://CRAN.R-project.org/package=rvest
Wickham H (2022). stringr: Simple, Consistent Wrappers for Common String Operations. https://CRAN.R-project.org/package=stringr
Wickham H (2016). ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York. ISBN 978-3-319-24277-4. https://CRAN.R-project.org/package=ggplot2
R Core Team (2013). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0.http://www.R-project.org/
Acknowledgements
This package was developed as part of an assessment for 2022 BCB410H:
Applied Bioinformatics course at the University of Toronto, Toronto,
CANADA. AbunRNA welcomes issues, enhancement requests, and other
contributions. To submit an issue, use the GitHub issues.