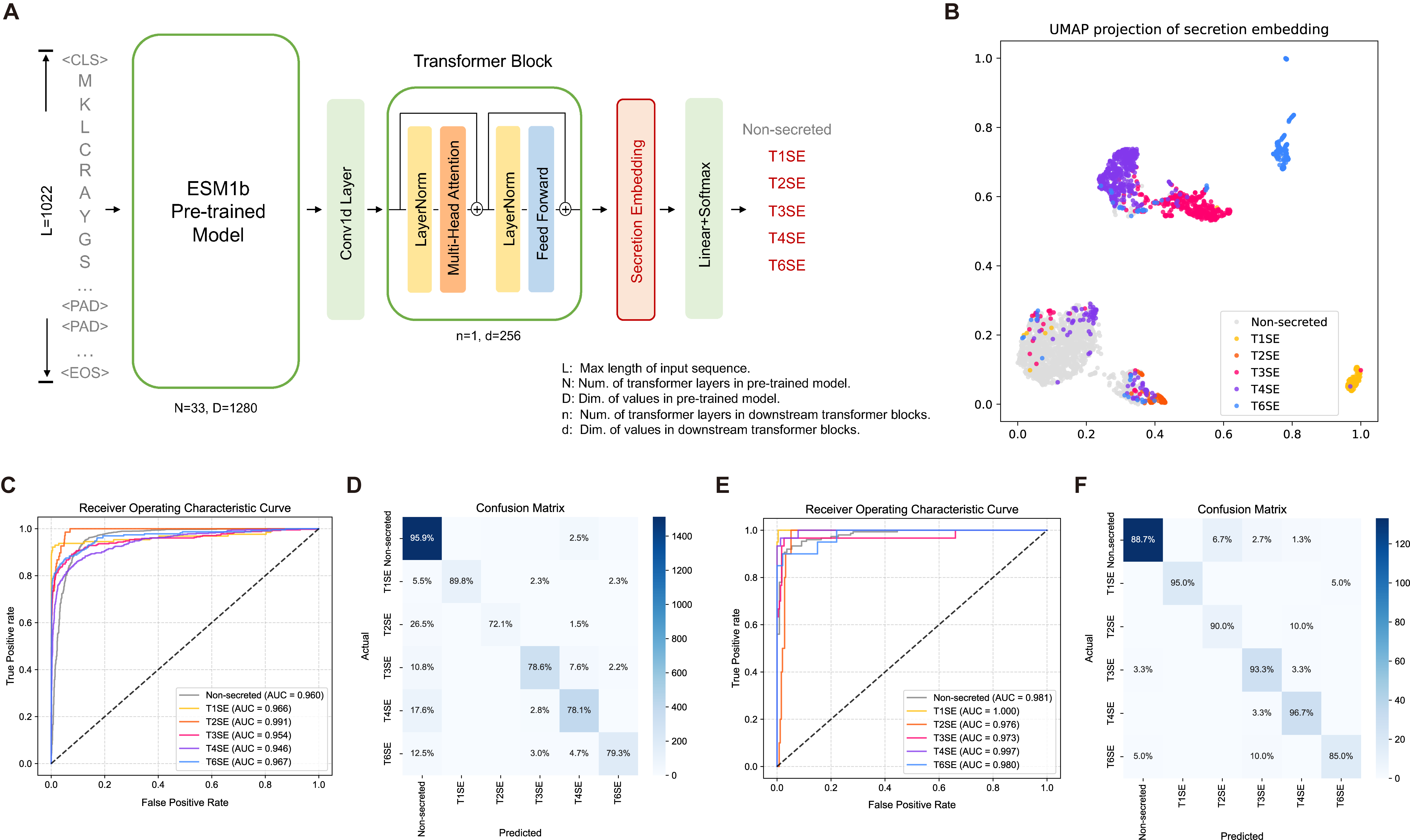
Implementation of Secretion-specific Transformer model used in secretion protein prediction in Gram-negative bacteria. DeepSecE achieves state-of-the-art performance in multi-class prediction leveraging the power of pre-trained protein language model ESM-1b. An additional transformer layer enhances the understanding of secreted patterns. It also provides a rapid pipeline to identify type I-IV and VI secretion systems with corresponding substrate proteins.
We choose various model architecture with different pre-trained models and training strategies, and evalute their model capacity on cross-validation and independent testing. Performance metrics are reported in the table.
| Pre-trained Model | Strategy | ACC | F1 | AUPRC | |||
|---|---|---|---|---|---|---|---|
| Valid | Test | Valid | Test | Valid | Test | ||
| / | PSSM+CNN | 0.799 | 0.822 | 0.712 | 0.724 | 0.752 | 0.774 |
| TAPEBert | Linear probing | 0.816 | 0.838 | 0.764 | 0.770 | 0.802 | 0.822 |
| ESM-1b | XGBoost | 0.869 | 0.887 | 0.809 | 0.816 | 0.865 | 0.872 |
| ESM-1b | Linear probing | 0.876 | 0.870 | 0.841 | 0.810 | 0.880 | 0.871 |
| ESM-1b | Fine-tuning | 0.878 | 0.850 | 0.846 | 0.808 | 0.887 | 0.883 |
| ESM-1b | Secretion-specific transformer | 0.883 | 0.898 | 0.848 | 0.849 | 0.892 | 0.885 |
- python==3.9.7
- torch==1.10.2
- biopython==1.79
- einops==0.4.1
- fair-esm>=0.4.0
- tqdm==4.64.0
- numpy==1.21.2
- scikit-learn==0.23.2
- matplotlib==3.5.1
- seaborn==0.11.0
- tensorboardX==2.0
- umap-learn==0.5.3
- warmup-scheduler==0.3.2
While we have not tested with other versions, any reasonably recent versions of these requirements should work.
As a prerequisite, you must have PyTorch installed. It is recommended to create a new virtual environment for installation. For model training and prediction from seperate protein sequence(s), You can use this one-liner for installation.
pip install DeepSecE==0.1.0 # OR
pip install git+https://github.com/zhangyumeng1sjtu/DeepSecE.gitIf you want to plot the sequence attention, you should install package logomarker first.
pip install logomakerIf you want to predict secretion systems and substrate proteins, you should install macsyfinder and hmmer first. Meanwhile, you need to download the TXSS profiles from here, and decompress it into data directory.
pip install macsyfinder
conda install -c bioconda hmmer
cd data
wget https://tool2-mml.sjtu.edu.cn/DeepSecE/TXSS_profiles.tar.gz
tar -zxvf TXSS_profiles.tar.gzThe weights of DeepSecE model can be downloaded from https://tool2-mml.sjtu.edu.cn/DeepSecE/checkpoint.pt.
You can train the DeepSecE model by running train.py or scripts/kfold_train.sh for cross-validation.
for i in {0..4}
do
python train.py --model effectortransformer \
--data_dir data \
--batch_size 32 \
--lr 5e-5 \
--weight_decay 4e-5 \
--dropout_rate 0.4 \
--num_layers 1 \
--num_heads 4 \
--warm_epochs 1 \
--patience 5 \
--lr_scheduler cosine \
--lr_decay_steps 30 \
--kfold 5 \
--fold_num $i \
--log_dir runs/attempt_cv
doneParameters:
--modeltrain a transformer or finetune a ESM-1b model.--data_dirdirectory that stores training data (default: ./data).--num_layersnumbers of trainable transformer layer. (default: 1)--num_headsnumbers of attention heads in secretion-specific transformer (default: 4).--patiencepatience for early stopping used in training.--lr_schedularlearning rate schedular [step, consine].--log_dirdirectory that stores training outputs (default: logs).
You can predict your interested type of secreted proteins only or predict secretion systems and corresponding substrate proteins from scratch.
python predict.py --fasta_path examples/Test.fasta \
--model_location [path to model weights] \
--secretion_systems [I II III IV VI] \
--out_dir examples [--save_attn --no_cuda]Parameters:
--fasta_pathpath to the input protein FASTA file.--model_locationpath to the model weights (download from here).--secretion_systemstype(s) of secretion system to predict (default: I II III IV VI).--out_dirdirectory that stores prediction outputs.--save_attnadd to save sequence attention of secreted protein.--no_cudaadd when CUDA is not available.
Note: Make sure the input file is ordered protein sequences coded in a bacterial genome.
python predict_genome.py --fasta_path examples/NC_002516.2_protein.fasta \
--model_location [path to model weights] \
--data_dir data \
--out_dir examples/NC_002516.2 [--save_attn --no_cuda]Parameters:
--fasta_pathpath to the input protein FASTA file.--model_locationpath to the model weights (download from here).--data_dirdirectory that stores TXSS profiles (download from here).--out_dirdirectory that stores prediction outputs.--save_attnadd to save sequence attention of secreted protein.--no_cudaadd when CUDA is not available.
It takes about 5 minutes to predict secreted proteins from a bacterial genome containing 3000 proteion coding sequences on a NVIDIA GeForce RTX 2080 Super GPU.
If you save the attention output of the putative secreted proteins (add --save_attn), you can run python scripts/plot_attention.py [directory of prediction output] to plot the saliency map from attention, and infer potentially import regions related to protein secretion.
Please contact Yumeng Zhang at zhangyumeng1@sjtu.edu.cn for questions.