The gtsummary package for R produces beautiful, customizable, publication-ready tables to summarize statistical models. Results from several models are presented side-by-side, with uncertainty estimates in parentheses (or brackets) underneath coefficient estimates.
Here are a few benefits of gtsummary over some alternative packages:
- html, rtf, and LaTeX output
- Excellent integration with:
- RStudio: When users type
gtsummary(models), the summary table immediately appears in RStudio's Viewer window. knitr: Dynamic document generation FTW.- Designed from the ground up with the
tidyparadigm in mind.
- RStudio: When users type
- Endlessly customizable tables, thanks to the power of the
gtpackage.- In the next section of this README, you will find tables with colored cells, weird text, spanning column labels, row groups, titles and subtitles, footnotes, significance stars, etc.
gtsummaryuses thebroompackage to extract information from model objects. This means thatgtsummarysupports dozens of model types out of the box. Most importantly,broomalready has a large community of users, and wheneverbroomimproves,gtsummaryimproves.- By using the
broomandgtpackage for key operations,gtsummaryhas a massively simplified codebase. This should improve long term code maintainability, and allow contributors to participate through GitHub. gtsummaryis developed with unit tests.
- Installation
- Using gtsummary
- Preliminaries
- Simple table
- SE, p, t, CI
- Output formats
- Titles and subtitles
- Group columns (spanning labels)
- Notes
- Rename, reorder, and subset coefficients
- Rename, reorder, and subset goodness-of-fit statistics
- Stars
- Digits, rounding, exponential notation
- Styles and colors
- Fancy text with markdown: bold, italics, etc.
- Complex table
- Power users
- Alternative summary table packages for R
The gt and gtsummary packages are not available on CRAN yet. You can install them from github:
library(remotes)
remotes::install_github('rstudio/gt')
remotes::install_github('vincentarelbundock/gtsummary')Make sure you also install tidyverse, as gtsummary depends on a lot of its packages (e.g., stringr, dplyr, tidyr, purrr):
install.packages('tidyverse')Load packages and download some data from the RDatasets repository. Then, estimate 5 different models and store them in a named list. The name of each model in that list will be used as a column label:
library(gt)
library(MASS)
library(gtsummary)
url <- 'https://vincentarelbundock.github.io/Rdatasets/csv/HistData/Guerry.csv'
dat <- read.csv(url)
dat$Clergy <- ifelse(dat$Clergy > 40, 1, 0) # binary variable for logit model
models <- list()
models[['OLS 1']] <- lm(Literacy ~ Crime_prop + Infants, dat)
models[['NBin 1']] <- glm.nb(Literacy ~ Crime_prop + Donations, dat)
models[['OLS 2']] <- lm(Desertion ~ Crime_prop + Infants, dat)
models[['NBin 2']] <- glm.nb(Desertion ~ Crime_prop + Donations, dat)
models[['Logit 1']] <- glm(Clergy ~ Crime_prop + Infants, dat, family = binomial())Produce a simple table.
gtsummary(models)RStudio will render this automatically as an html table. If you do not use RStudio, read the next section to learn how to save to file.
Of course, gtsummary can also summarize single models:
mod <- lm(Clergy ~ Crime_prop, data = dat)
gtsummary(mod)To save a table to file, use the filename argument. gtsummary guesses the output format based on the filename extension. The supported extensions are: .tex, .rtf, .html (ASCII/Text tables coming soon).
gtsummary(models, filename = 'table.tex')
gtsummary(models, filename = 'table.rtf')
gtsummary(models, filename = 'table.html')If filename is not specified, gtsummary returns a gt object which can be further customized and rendered by the relevant functions in the gt package, such as as_raw_html, as_latex, or as_rtf. RStudio renders the html version of this object automatically.
gtsummary prints an uncertainty estimate in parentheses below the corresponding coefficient estimate. The statistic argument must be a string which is equal to conf.int or to one of the columns produced by the broom::tidy function. When using conf.int, users can specify a confidence level with the conf_level argument.
gtsummary(models, statistic = 'std.error')
gtsummary(models, statistic = 'p.value')
gtsummary(models, statistic = 'statistic')
gtsummary(models, statistic = 'conf.int', conf_level = .99)You can add titles and subtitles to your table as follows:
gtsummary(models,
title = 'This is a title for my table.',
subtitle = 'And this is the subtitle.')Add notes to the bottom of your table:
gtsummary(models,
notes = list('Text of the first note.',
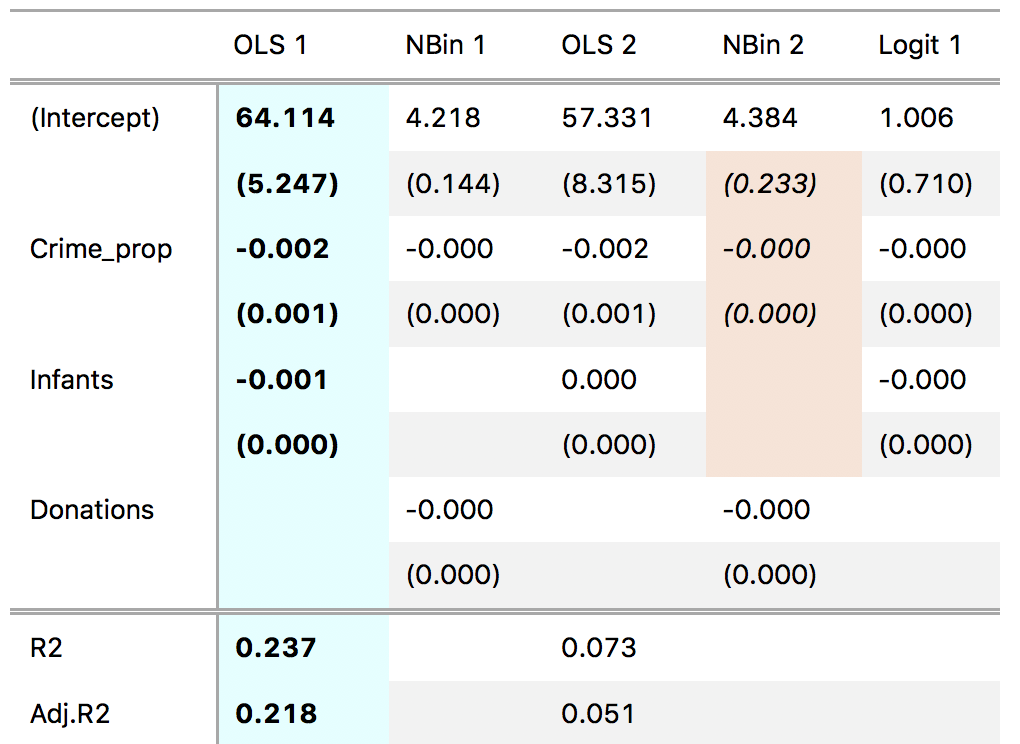
'Text of the second note.'))The coef_map argument is a named vector which allows users to rename, reorder, and subset coefficient estimates. Values of this vector correspond to the "clean" variable name. Names of this vector correspond to the "raw" variable name. The table will be sorted in the order in which terms are presented in coef_map. Coefficients which are not included in coef_map will be excluded from the table.
cm <- c('Crime_prop' = 'Crime / Population',
'Donations' = 'Donations',
'(Intercept)' = 'Constant')
gtsummary(models, coef_map = cm)An alternative mechanism to subset coefficients is to use the coef_omit argument. This string is a regular expression which will be fed to stringr::str_detect to detect the variable names which should be excluded from the table.
gtsummary(models, coef_omit = 'Intercept|Donation')gof_omit is a regular expression which will be fed to stringr::str_detect to detect the names of the statistics which should be excluded from the table.
gtsummary(models, gof_omit = 'DF|Deviance')Create spanning labels to group models (columns):
gtsummary(models) %>%
gt::tab_spanner(label = 'Literacy', columns = c('OLS 1', 'NBin 1')) %>%
gt::tab_spanner(label = 'Desertion', columns = c('OLS 2', 'NBin 2')) %>%
gt::tab_spanner(label = 'Clergy', columns = 'Logit 1')Some people like to add "stars" to their model summary tables to mark statistical significance. The stars argument can take three types of input:
NULLomits any stars or special marks (default)TRUEuses these default values:* p < 0.1, ** p < 0.05, *** p < 0.01- Named numeric vector for custom stars.
gtsummary(models)
gtsummary(models, stars = TRUE)
gtsummary(models, stars = c('+' = .1, '*' = .01)) The fmt argument defines how numeric values are rounded and presented in the table. This argument follows the sprintf C-library standard. For example,
%.3fwill keep 3 digits after the decimal point, including trailing zeros.%.5fwill keep 5 digits after the decimal point, including trailing zeros.- Changing the
ffor anewill use the exponential decimal representation.
Most users will just modify the 3 in %.3f, but this is a very powerful system, and all users are encouraged to read the details: ?sprintf
gtsummary(models, fmt = '%.7f')The power of the gt package makes gtsummary tables endlessly customizable. For instance, we can color columns and cells, and present values in bold or italics:
gtsummary(models) %>%
tab_style(style = cells_styles(bkgd_color = "lightcyan",
text_weight = "bold"),
locations = cells_data(columns = vars(`OLS 1`))) %>%
tab_style(style = cells_styles(bkgd_color = "#F9E3D6",
text_style = "italic"),
locations = cells_data(columns = vars(`NBin 2`),
rows = 2:6))Thanks to gt, gtsummary accepts markdown indications for emphasis and more:
gtsummary(models,
title = md('This is a **bolded series of words.**'),
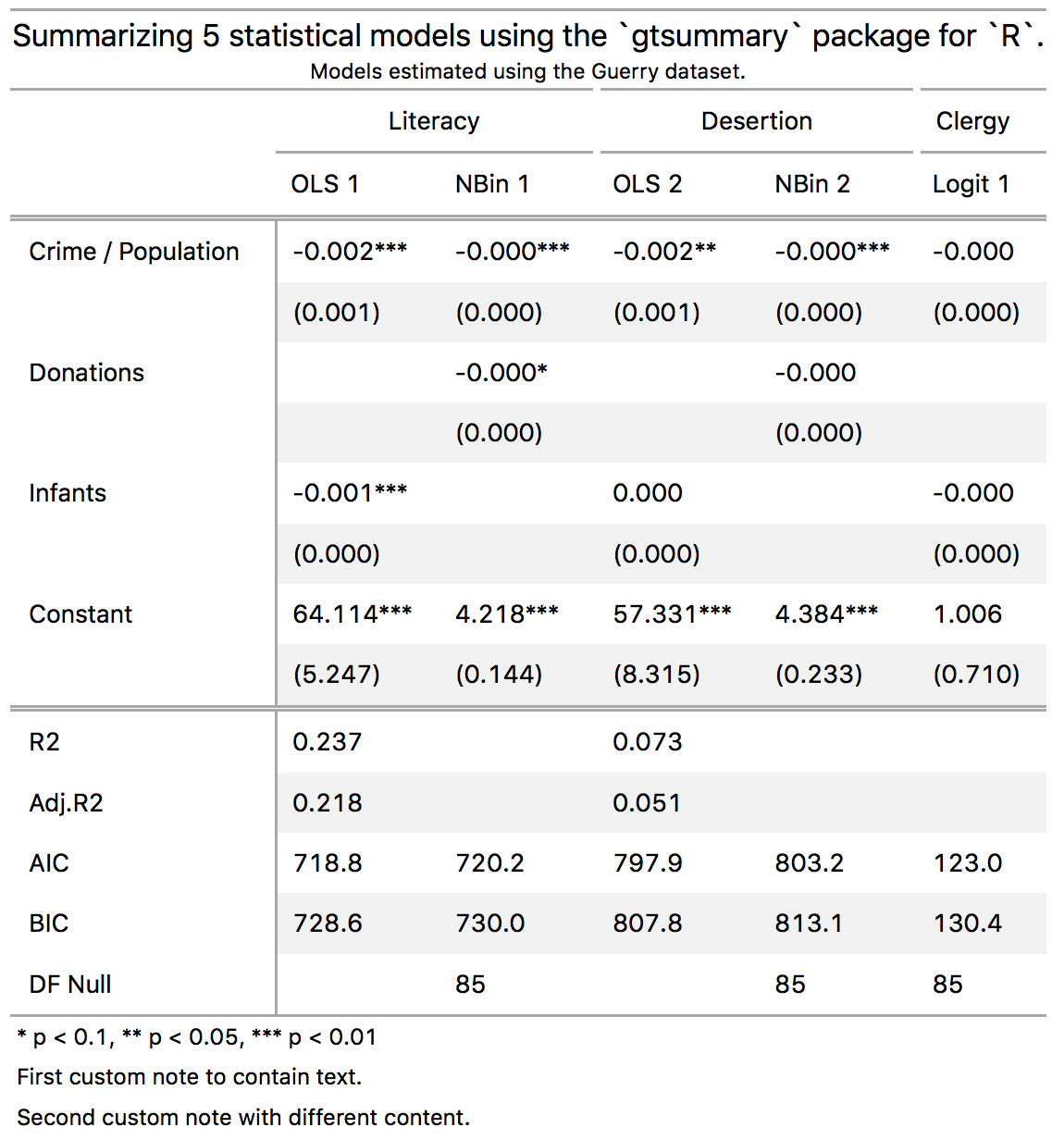
notes = list(md('And an *emphasized note*.')))This is the code I used to generate the "complex" table posted at the top of this README.
cm <- c('Crime_prop' = 'Crime / Population',
'Donations' = 'Donations',
'Infants' = 'Infants',
'(Intercept)' = 'Constant')
gtsummary(models,
coef_map = cm,
stars = TRUE,
gof_omit = "Statistics|^p$|Deviance|Resid|Sigma|Log.Lik|^DF$",
title = 'Summarizing 5 statistical models using the `gtsummary` package for `R`.',
subtitle = 'Models estimated using the Guerry dataset.',
notes = c('First custom note to contain text.',
'Second custom note with different content.')) %>%
# add spanning labels
gt::tab_spanner(label = 'Literacy', columns = c('OLS 1', 'NBin 1')) %>%
gt::tab_spanner(label = 'Desertion', columns = c('OLS 2', 'NBin 2')) %>%
gt::tab_spanner(label = 'Clergy', columns = 'Logit 1')The gt package allows a bunch more customization and styling. Power users can use gtsummary's extract function to produce a tibble which can easily be fed into gt.
> gtsummary::extract(models)
# A tibble: 21 x 8
group term statistic `OLS 1` `NBin 1` `OLS 2` `NBin 2` `Logit 1`
<chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
1 estimates (Intercept) estimate 64.114 4.218 57.331 4.384 1.006
2 estimates (Intercept) statistic (5.247) (0.144) (8.315) (0.233) (0.710)
3 estimates Crime_prop estimate -0.002 -0.000 -0.002 -0.000 -0.000
4 estimates Crime_prop statistic (0.001) (0.000) (0.001) (0.000) (0.000)
5 estimates Infants estimate -0.001 "" 0.000 "" -0.000
6 estimates Infants statistic (0.000) "" (0.000) "" (0.000)
7 estimates Donations estimate "" -0.000 "" -0.000 ""
8 estimates Donations statistic "" (0.000) "" (0.000) ""
9 gof R2 "" 0.237 "" 0.073 "" ""
10 gof Adj.R2 "" 0.218 "" 0.051 "" ""
# … with 11 more rowsThere are several excellent alternative summary table packages for R: