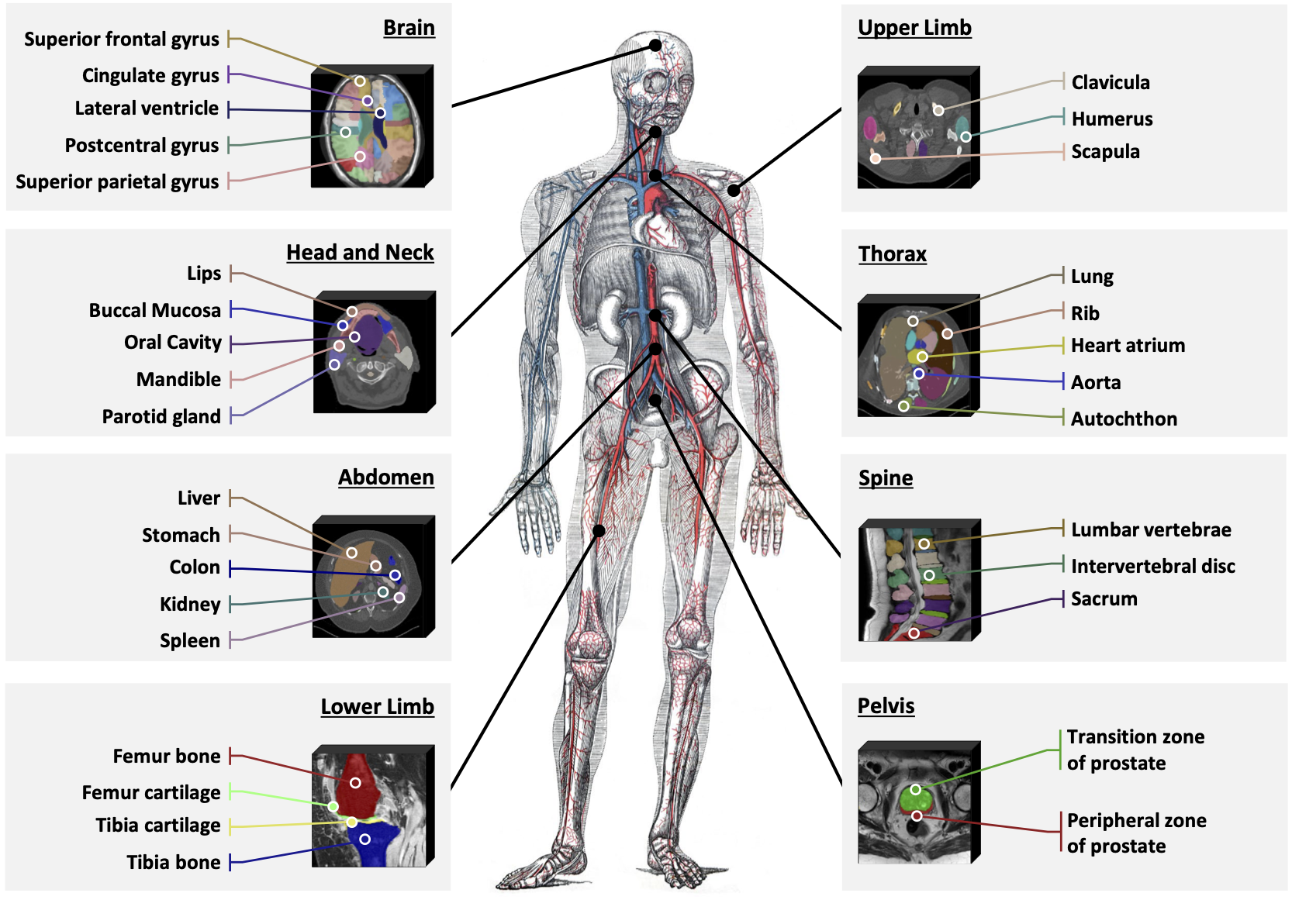
This is the official repository to build SAT-DS, a medical data collection of 72 public segmentation datasets, contains over 22K 3D images, 302K segmentation masks and 497 classes from 3 different modalities (MRI, CT, PET) and 8 human body regions. 🚀
Based on this data collection, we build an universal segmentation model for 3D radiology scans driven by text prompts (check this repo and our paper).
The data collection will continuously growing, stay tuned!
🎉 To save your time from downloading and preprocess so many datasets, we offer shortcut download links of 42/72 datasets in SAT-DS, which allow re-attribution with licenses such as CC BY-SA. Find them in dropbox.
All these datasets are preprocessed and packaged by us for your convenience, ready for immediate use upon download and extraction. Download the datasets you need and unzip them in data/nii, these datasets can be used immediately with the paired jsonl files in data/jsonl, check Step 3 below for how to use them. Note that we respect and adhere to the licenses of all the datasets, if we incorrectly reattribute any of them, please contact us.
- Collect as many public datasets as possible for 3D medical segmentation, and compile their basic information;
- Check and normalize image scans in each dataset, including orientation, spacing and intensity;
- Check, standardize, and merge the label names for categories in each dataset;
- Carefully split each dataset into train and test set by the patient id.
- (Step 1) Access to each dataset in SAT-DS.
- (Step 2) Code to preprocess samples in each dataset.
- (Shortcut to skip Step 1 and 2) Access to preprocessed and packaged datasets that can be used immediately.
- (Step 3) Code to load samples with normalized image, standardized class names from each dataset.
- (Step 3) Code to visualize and check the samples.
- (Step 4) Code to prepare the train and evaluation data for SAT in required format.
- (Step 5) Code to split the dataset into train and test in consistent with SAT.
- (Follow step 1~3) Preprocess and unfied a large-scale and comprehensive 3D medical segmentation data collection, suitable to train or finetune universal segmentation models like SAM2.
- (Follow step 1~6) Prepare the training and test data in required format for SAT.
Check our paper "One Model to Rule them All: Towards Universal Segmentation for Medical Images with Text Prompts" for more details.
This is the detailed list of all the datasets and their official download links. Their citation information can be found in citation.bib .
As a shortcut, we preprocess, package and re-attribute some of them for your convenient use. Download them here.
For each dataset, we need to find all the image and mask pairs, and another 5 basic information: dataset name, modality, label name, patient ids (to split train-test set) and official split (if provided).
In processor.py, we customize the process procedure for each dataset, to generate a jsonl file including these information for each sample.
Take AbdomenCT1K for instance, you need to run the following command:
python processor.py \
--dataset_name AbdomenCT1K \
--root_path 'SAT-DS/data/nii/AbdomenCT-1K' \
--jsonl_dir 'SAT-DS/data/jsonl'
root_path should be where you download and place the data, jsonl_dir should be where you plan to place the jsonl files.
dataset_name and the name in the table might not be exactly the same. For specific details, please refer to each process function in processor.py.
After process, each sample in jsonl files would be like:
{
'image' :"SAT-DS/data/nii/AbdomenCT-1K/Images/Case_00558_0000.nii.gz",
'mask': "SAT-DS/data/nii/AbdomenCT-1K/Masks/Case_00558.nii.gz",
'label': ["liver", "kidney", "spleen", "pancreas"],
'modality': 'CT',
'dataset': 'AbdomenCT1K,
'official_split': 'unknown',
'patient_id': 'Case_00558_0000.nii.gz',
}
Note that in this step, we may convert the image and mask into new nifiti files for some datasets, such as TotalSegmentator and so on. So it may take some time.
We offer shortcut download links of 42 datasets in dropbox. All these datasets are preprocessed and packaged in advance. Download the datasets you need and unzip them in data/nii, each dataset is paired with a jsonl file in data/jsonl.
With the generated jsonl file, a dataset is now ready to be used.
However, when mixing all the datasets to train a universal segmentation model, we need to apply normalization on the image intensity, orientation, spacing across all the datasets, and adjust labels if necessary.
We realize this by customizing the load script for each dataset in loader.py, this is a simple demo how to use it in your code:
from loader import Loader_Wrapper
loader = Loader_Wrapper()
# load samples from jsonl
with open('SAT-DS/data/jsonl', 'r') as f:
lines = f.readlines()
data = [json.loads(line) for line in lines]
# load a sample
for sample in data:
batch = getattr(loader, func_name)(sample)
img_tensor, mc_mask, text_ls, modality, image_path, mask_path = batch
For each sample, whatever the dataset it comes from, the loader will give output in a normalized format:
img_tensor # tensor with shape (1, H, W, D)
mc_mask # binary tensor with shape (N, H, W, D), one channel for each class;
text_ls # a list of N class name;
modality # MRI, CT or PET;
image_path # path to the loaded mask file;
mask_path # path to the loaded imag file;
text_ls may be different from the label you see in the input jsonl file.
Here is an case where we merge left kidney' and right kidneyfor a new labelkidney` when loading examples from CHAOS_MRI:
kidney = mask[1] + mask[2]
mask = torch.cat((mask, kidney.unsqueeze(0)), dim=0)
labels.append("kidney")
And here is another case where we adjust the annotation of kidney by integrating the annotation of kidney tumor and kidney cyst:
mc_masks[0] += mc_masks[1]
mc_masks[0] += mc_masks[2]
We also offer the shortcut to visualize and check any sample in any dataset after normalization. For example, to visualize the first sample in AbdomenCT1K.jsonl, just run the following command:
python loader.py \
--visualization_dir 'SAT-DS/data/visualization' \
--path2jsonl 'SAT-DS/data/jsonl/AbdomenCT1K.jsonl' \
--i 0
For convenience, before training SAT, we normalize all the data according to step 3, and convert the images and segmentation masks to npy files. If you try to use our training code, run this command for each dataset:
python convert_to_npy.py \
--jsonl2load 'SAT-DS/data/jsonl/AbdomenCT1K.jsonl' \
--jsonl2save 'SAT-DS/data/jsonl/AbdomenCT1K.jsonl'
The converted npy files will be saved in preprocessed_npy/dataset_name, and some new information will be added to the jsonl file for connivence to load the npy files.
We offer the train-test split used in our paper for each dataset in json files. To follow our split and benchmark your method, simply run this command:
python train_test_split.py \
--jsonl2split 'SAT-DS/data/jsonl/AbdomenCT1K.jsonl' \
--train_jsonl 'SAT-DS/data/trainset_jsonl/AbdomenCT1K.jsonl' \
--test_jsonl 'SAT-DS/data/testset_jsonl/AbdomenCT1K.jsonl' \
--split_json 'SAT-DS/data/split_json/AbdomenCT1K.json'
This will split the jsonl file into train and test.
Or, if you want to re-split them, just customize your split by identifying the patient_id in the json file (patient_id of each sample can be found in jsonl file of each dataset):
{'train':['train_patient_id1', ...], 'test':['test_patient_id1', ...]}
You may want to customize the dataset collection in training your model, simply merge the train jsonls of the data you want to involve. For example, merge the jsonls for all the 72 datasets into train.jsonl, and you can use them together to train SAT, using our training code in this repo.
Similarly, you can customize a benchmark with arbitrary datasets you want by merging the test jsonls.
If you use this code for your research or project, please cite:
@arxiv{zhao2023model,
title={One Model to Rule them All: Towards Universal Segmentation for Medical Images with Text Prompt},
author={Ziheng Zhao and Yao Zhang and Chaoyi Wu and Xiaoman Zhang and Ya Zhang and Yanfeng Wang and Weidi Xie},
year={2023},
journal={arXiv preprint arXiv:2312.17183},
}
And if you use any of these datasets in SAT-DS, please cite the corresponding papers. A summerized citation information can be found in citation.bib .