This is the repository for the BIOF501A Final Project .
COVID-19 is a global pandemic and represents one of the toughest health challenges in 2020 [1]. As cases and mortality have been rapidly increasing over the past months, the need of rapid information regarding COVID-19 sequences for scientists and clinicians have increased dramatically [1]. While existing COVID-19 pipelines for sequencing have been developed [2]. To my knowledge, few pipelines have utilized the Phylogenetic Assignment of Named Global Outbreak LINeages (PANGOLIN) software, which is a software that can rapidly detect the lineage from a given fasta file. Overall, 5 FASTQ samples submitted by the Delaware Public Health Lab to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) have been identified and will be used to demonstrate the validity of this pipeline. As the US is seeing unprecendented rise in the number of COVID cases, rapid and urgent responses must be done as quickly as possible, and this pipeline offers an expedient manner of collecting lineage. Theoretically, this may be relevant clinically relevant as physicians might notice that a cluster of cases have a common lineage, which may inform they of either a superspread event occuring or whether distinct, separate local outbreaks are occuring. While currently only 1 sample is processed, more can be used in the imaginable future if this pipeline is further developed. Based on previous literature which suggest that type A and type C are found in significant proportions in Europeans and Americans, I hypothesize that the majority of samples will be either type A or type B lineage[3]. Note that for this analysis, only paired-end reads data was used.
Currently, git and Miniconda are required to run this program. It will be implicitly assumed that users have this already installed.
Before getting started, note that the reference sequence has already been provided for convenience, and can be found here. Briefly, the pipeline was built with Snakemake and is split into the following steps:
- Extracting FASTQ reads through the
sra-toolkit. - Mapping the reads to the reference sequence using
minimap2. - Creating fasta file through
samtools. - Running
pangolinto generate a .csv file of the result. - Creating a histogram using
matplotlibof the lineages counts.
To clone the repository, run the following shell command:
git clone https://github.com/zhemingfan/biof501a-mbb659_jeremy_fan.gitOnce the repository has been downloaded, more effort needs to be done to get the full installation. Unfortunately, an environment.yml could not be provided as pangolin must be downloaded in a specific manner.
Start by following the normal pangolin installation:
- Clone the repository at a location of your choosing.
git clone https://github.com/cov-lineages/pangolin.git - Go into the
pangolinfolder.
cd pangolin- Create the
condaenvironment file.
conda env create -f environment.yml- Activate the
condaenvironment.
conda activate pangolin- Run setup.py the file to finalize installation
python setup.py installIn one chunk, this is:
git clone https://github.com/cov-lineages/pangolin.git
cd pangolin
conda env create -f environment.yml
conda activate pangolin
python setup.py installAfterwards, install a suite of tools from bioconda, conda-forge, and conda. Check yes to everything by clicking the "y" key.
conda install -c bioconda sra-tools samtools=1.9 openssl=1.0 bcftools seqtk
conda install -c conda-forge matplotlib
conda install pandasTo run the pipeline, go into the directory containing the Snakemake file, make sure the pangolin conda environment is activated and run the following:
snakemake --cores 1 The total expected running time should be at most 10 minutes (excluding data download, which may take the longer).
-
Extracting FASTQ reads through the
sra-toolkit. -fasterq-dump --split-files SRR12960723 -O data:fasterq-dumpis a faster way to extract fastq files from thesra-toolkit.--split-filesindicate split reads, and the-Oflag suggests output to a directory called data -afterwards, acatis run to append the two paired ends read files together. If successful, remove the individual fastq to save storage space -
Mapping the reads to the reference sequence using
minimap2. -minimap2is a [https://github.com/lh3/minimap2](general purpose alligner), but largely maintains much of the performance asbwa-memfor short reads. the-ax srflag is added to specify short reads.samtoolsis then called to sort the data by leftmost coordinates -for the future, if ever long reads are added to this pipeline,minimap2would make an ideal choice -
Creating fasta file through
samtools. -the following steps are adapted from an answer from https://www.biostars.org/p/367626/. Briefly,mpileupproduces "pileup" textual format from an alignment. then an eventual "consensus" of the most frequently occuring base at each spot is generated throughbcftools callandvcfutils.pl vcf2fq. This practically does the same asbcftools consensus. -
Running
pangolinto generate a .csv file of the result. -pangolinis fairly self contained package - basically a multinomial logistic regression model built using 30,000 SARS-CoV-2 sequences from GISAID -
Creating a histogram using
matplotlibof the lineages counts. -pandasis used to read the csv file -matplotlibhas a built in histogram feature that can be called with
The first input file provided in the data folder is a sequence of the Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome. This file follows traditional FASTA format (one line header starting with ">" following a sequence of nucleotides in the second line). Five additional fastq files can be found from SRA, for instance one of the read file is here. The accession numbers are: SRR12960723, SRR12960724, SRR12960725, SRR12960726, SRR12960727. All of the read files are standard, and are paired-ends, thus accounting for why later a cat command will be run to merge them.
After running the pipeline, the final directory should look something similar to below (note hidden files are excluded):
To be noted:
- combined.fasta is a fasta file with the combined fasta from all the samples.
- newcombined.fasta is another file except all the headers are changed to allow analysis (previously all headers were the same)
- A series of .bam files such as SRR12960723.alignments.sorted.bam are the results of the allignment between reads and the reference genome provided.
- Within the results folder, there will be two new files
- lineage_report.csv which provides a report on the samples and the lineage they were called
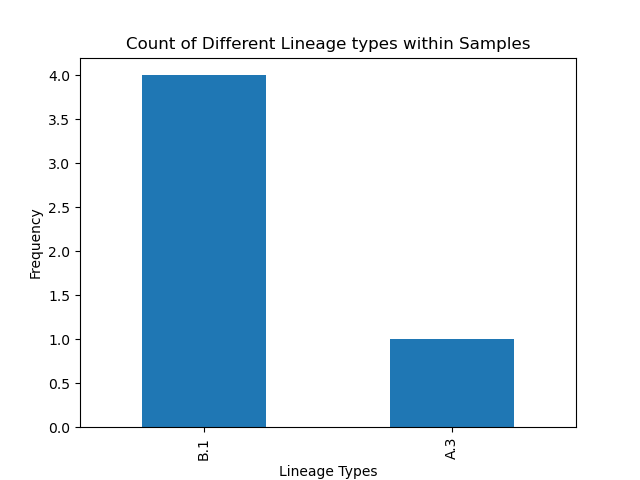
- covid_histogram.png which provides a visual perspective of the same data in the lineage report
Overall, while lineages A and C were anticipated, most samples were classified as lineage B.1. In other manuscripts, however, the B.1 lineage has also spread to North America, and was noted to start in New York City [4][5]. This specific lineage contains a specific mutation (D614G) within the Spike protein that has been been linked with higher contagion [6]. Newer reports seem to indicate that both B.1 and A.3 have also been commonly observed across the United States [7]. However, as the sample size was only limited to 5, it is difficult to come with definitive conclusions about which lineages are most common in Delaware. As more data is later acquired, more evidence can be brought up and perhaps targeted therapies at specific mutations in specific lineages may arise.
| Team Member | Degree | PI | Hobbies |
|---|---|---|---|
| Jeremy Fan | Bioinformatics | Steven Jones | Annoying my roommate by cooking instant noodles at 3 AM |
[1] Lee, M. (2020). Clinical Characteristics Of Early Noncritical Hospitalized Patients With Coronavirus Disease 2019 (Covid-19): A Single-Center Retrospective Study In New York City. doi:10.26226/morressier.5ebc261fffea6f735881a237
[2] Nasir, Jalees A., Robert A. Kozak, Patryk Aftanas, Amogelang R. Raphenya, Kendrick M. Smith, Finlay Maguire, Hassaan Maan et al. "A Comparison of Whole Genome Sequencing of SARS-CoV-2 Using Amplicon-Based Sequencing, Random Hexamers, and Bait Capture." Viruses 12, no. 8 (2020): 895. https://doi.org/10.3390/v12080895
[3] Peter Forster, Lucy Forster, Colin Renfrew, Michael Forster. "Phylogenetic network analysis of SARS-CoV-2 genomes". Proceedings of the National Academy of Sciences Apr 2020, 117 (17) 9241-9243; DOI: 10.1073/pnas.2004999117
[4] Worobey M, Pekar J, Larsen BB, et al. The emergence of SARS-CoV-2 in Europe and the US. Preprint. bioRxiv. 2020;2020.05.21.109322. Published 2020 May 23. doi:10.1101/2020.05.21.109322
[5] Maurano M. T., Ramaswami S., Westby G., Zappile P., Dimartino D., Shen G., Feng X., Ribeiro-dos-Santos A. M., Vulpescu N. A., Black M., Hogan M., Marier C., Meyn P., Zhang Y., Cadley J., Ordonez R., Luther R., Huang E., Guzman E., Serrano A., Belovarac B., Gindin T., Lytle A., Pinnell J., Vougiouklakis T., Boytard L., Chen J., Lin L. H., Rapkiewicz A., Raabe V., Samanovic-Golden M. I., Jour G., Osman I., Aguero-Rosenfeld M., Mulligan M. J., Cotzia P., Snuderl M., Heguy A., Sequencing identifies multiple, early introductions of SARS-CoV2 to New York City Region. medRxiv (2020), , doi:10.1101/2020.04.15.20064931
[6] Korber B., Fischer W. M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Foley B., Giorgi E. E., Bhattacharya T., Parker M. D., Partridge D. G., Evans C. M., Freeman T. M., de Silva T. I., on behalf of the Sheffield COVID-19 Genomics Group, LaBranche C. C., Montefiori D. C., Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2. bioRxiv (2020), p. 2020.04.29.069054.
[7] Ladner JT, Larsen BB, Bowers JR, Hepp CM, Bolyen E, Folkerts M, Sheridan K, Pfeiffer A, Yaglom H, Lemmer D, Sahl JW, Kaelin EA, Maqsood R, Bokulich NA, Quirk G, Watts TD, Komatsu K, Waddell V, Lim ES, Caporaso JG, Engelthaler DM, Worobey M, Keim P. 2020. An early pandemic analysis of SARS-CoV-2 population structure and dynamics in Arizona. mBio 11:e02107-20. https://doi.org/10.1128/mBio.02107-20.