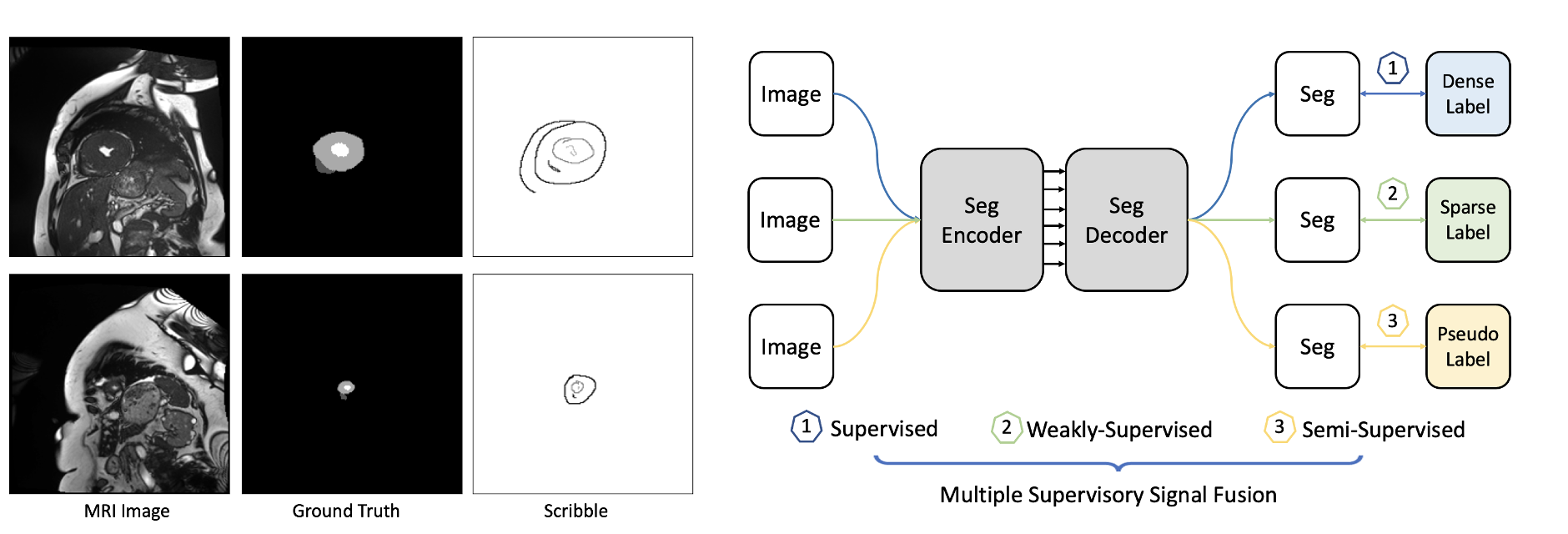
MixSegNet: Fusing multiple mixed-supervisory signals with multiple views of networks for mixed-supervised medical image segmentation
Deep learning has driven remarkable advancements in medical image segmentation. However, the requirement for comprehensive annotations poses a significant challenge due to the labor-intensive and expensive nature of expert annotation. Addressing this challenge, we introduce a fused limited-supervised learning strategy, MixSegNet, that synergistically harnesses the benefits of Fully-Supervised (FSL), Weakly-Supervised (WSL), and Semi-Supervised Learning (SSL). This approach enables the utilization of various data-efficient annotations for network training, promoting efficient medical image segmentation within realistic clinical scenarios.
- Pytorch
- Some basic python packages such as Numpy, Scikit-image, SimpleITK, Scipy ......
We use the ACDC dataset which you can find here Official. The pre-processed dataset i.e. scribble is with this GitHub Respository, and you can also simulate the scribble annotations with other dataset with the 'code/scribbles_generator.py' file.
- Clone the repo:
git clone https://github.com/ziyangwang007/MixSegNet.git
cd MixSegNet
- Train the model
cd code
python train_FuseSegNet.py
- Test the model
python test_2D_fully.py
For all other baseline methods, please check our work on Semi-Supervised Baseline Methods
and
@article{wang2024mixsegnet,
title={MixSegNet: Fusing multiple mixed-supervisory signals with multiple views of networks for mixed-supervised medical image segmentation},
author={Wang, Ziyang and Yang, Chen},
journal={Engineering Applications of Artificial Intelligence},
volume={133},
pages={108059},
year={2024},
publisher={Elsevier}
}