This repository is for "Graph-Skeleton: ~1% Nodes are Sufficient to Represent Billion-Scale Graph accepted by WWW2024.
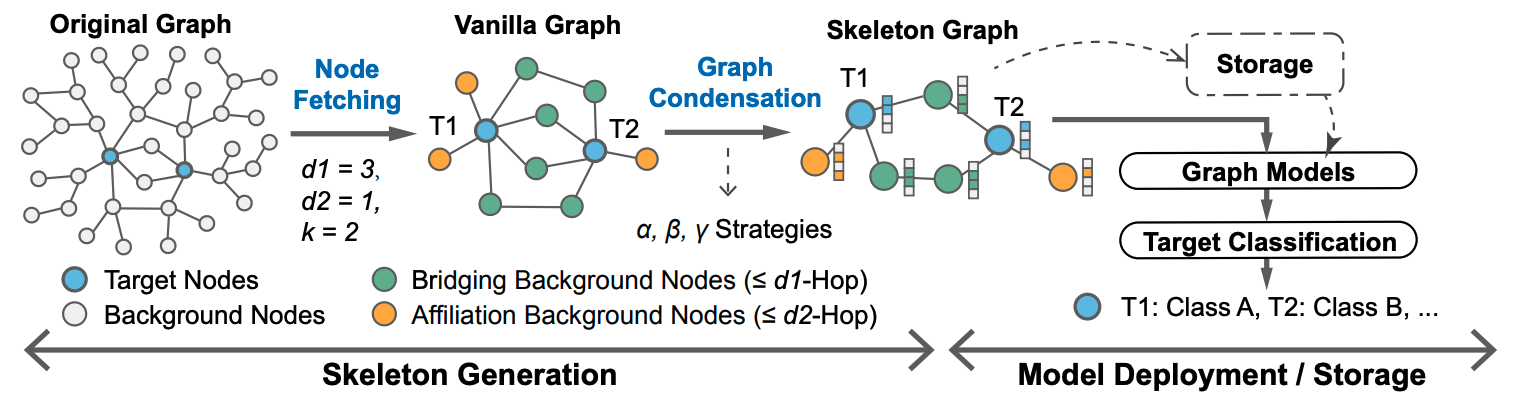
In this paper, we focus on a common challenge in graph mining applications: compressing the massive background nodes for a small part of target nodes classification, to tackle the storage and model deployment difficulties on very large graphs. The proposed Graph-Skeleton first fetches the essential background nodes under the guidance of structural connectivity and feature correlation, then condenses the information of background nodes with three different condensation strategies. The generated skeleton graph is highly informative and friendly for storage and graph model deployment.
Install libboost
sudo apt-get install libboost-all-devThe currently code demo is based on the dataset DGraph-Fin
. Please download the DGraphFin.zip and move it under directory datasets, unizp the dataset folder and organize as follows:
.
--datasets
└─DGraphFin
└─dgraphfin.npz
You can run the scripts in fold ./preliminary_exploration to implement the exploration studies (Section 2 [Empirical Analysis]) in our paper.
The edge cutting type is controlled by hyper-parameters --cut, which includes random cut (cut_random), cut T-T (tt), cut T-B (tb), cut B-B (bb).
cd preliminary_exploration
python dgraph/gnn_mini_batch.py --cut tt
python dgraph/gnn_mini_batch.py --cut cut_random --rd_ratio 0.5- Compile Graph-Skeleton
cd skeleton_compress
mkdir build
cd build
cmake ..
make
cd ..- Graph-Skeleton Generation
You can run the script ./skeleton_compress/skeleton_compression.py to generate skeleton graphs. Please note that in our original paper, hyper-parameters --d is set as [2,1], you can also modify the setting of d to change the node fetching distance.
By setting different values of hyper-parameters --cut, different strategies (i.e.,
python skeleton_compression.pyYou can run the scripts in fold ./skeleton_test to deploy the generated skeleton graphs on downstream GNNs for target nodes classification task.
cd skeleton_test/dgraph
python gnn_mini_batch.py --cut skeleton_gamma