A set of functions to analyse and plot the KEGG information using
tidygraph, ggraph and ggplot2.
The detailed documentation is
here using bookdown.
Using BiocManager:
BiocManager::install("ggkegg")Using devtools:
devtools::install_github("noriakis/ggkegg")library(ggkegg)
library(ggfx)
library(igraph)
library(tidygraph)
library(dplyr)
pathway("ko01100") |>
process_line() |>
highlight_module(module("M00021")) |>
highlight_module(module("M00338")) |>
ggraph(x=x, y=y) +
geom_node_point(size=1, aes(color=I(fgcolor),
filter=fgcolor!="none" & type!="line"))+
geom_edge_link0(width=0.1, aes(color=I(fgcolor),
filter=type=="line"& fgcolor!="none"))+
with_outer_glow(
geom_edge_link0(width=1,
aes(color=I(fgcolor),
filter=(M00021 | M00338))),
colour="red", expand=5
)+
with_outer_glow(
geom_node_point(size=1.5,
aes(color=I(fgcolor),
filter=(M00021 | M00338))),
colour="red", expand=5
)+
geom_node_text(size=2,
aes(x=x, y=y,
label=graphics_name,
filter=name=="path:ko00270"),
repel=TRUE, family="sans", bg.colour="white")+
theme_void()You can use your favorite geoms to annotate raw KEGG map combining the functions.
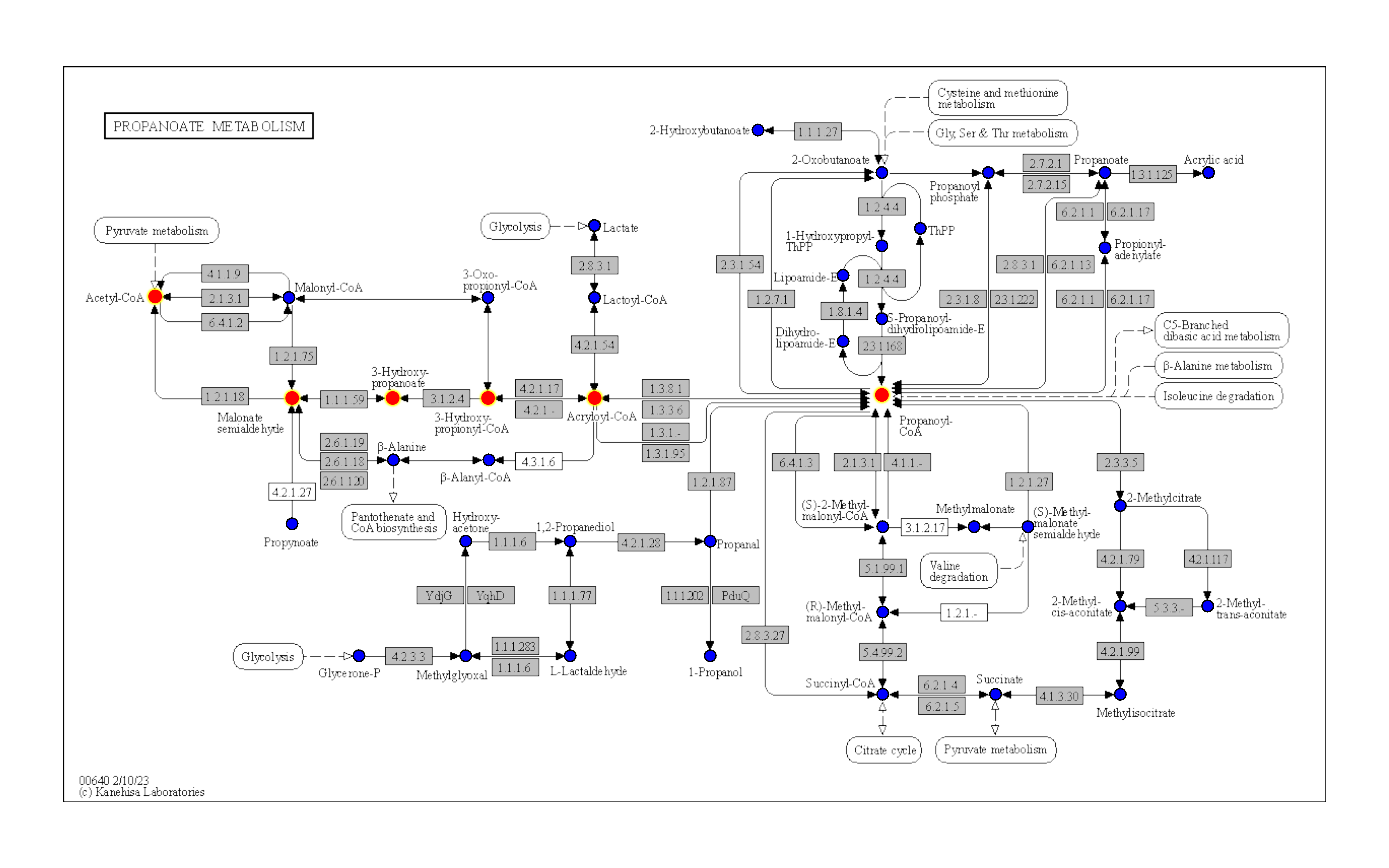
m <- module("M00013")
g <- pathway("ko00640") |> mutate(mod=highlight_set_nodes(m@reaction_components,how="all"))
gg <- ggraph(g, layout="manual", x=x, y=y)+
geom_node_rect(fill="grey",aes(filter=type=="ortholog"))+
overlay_raw_map("ko00640")+
geom_node_point(aes(filter=type=="compound"), shape=21, fill="blue", color="black", size=2)+
ggfx::with_outer_glow(geom_node_point(aes(filter=mod, x=x, y=y), color="red",size=2),
colour="yellow",expand=5)+
theme_void()
ggOr customize graphics based on ggraph.
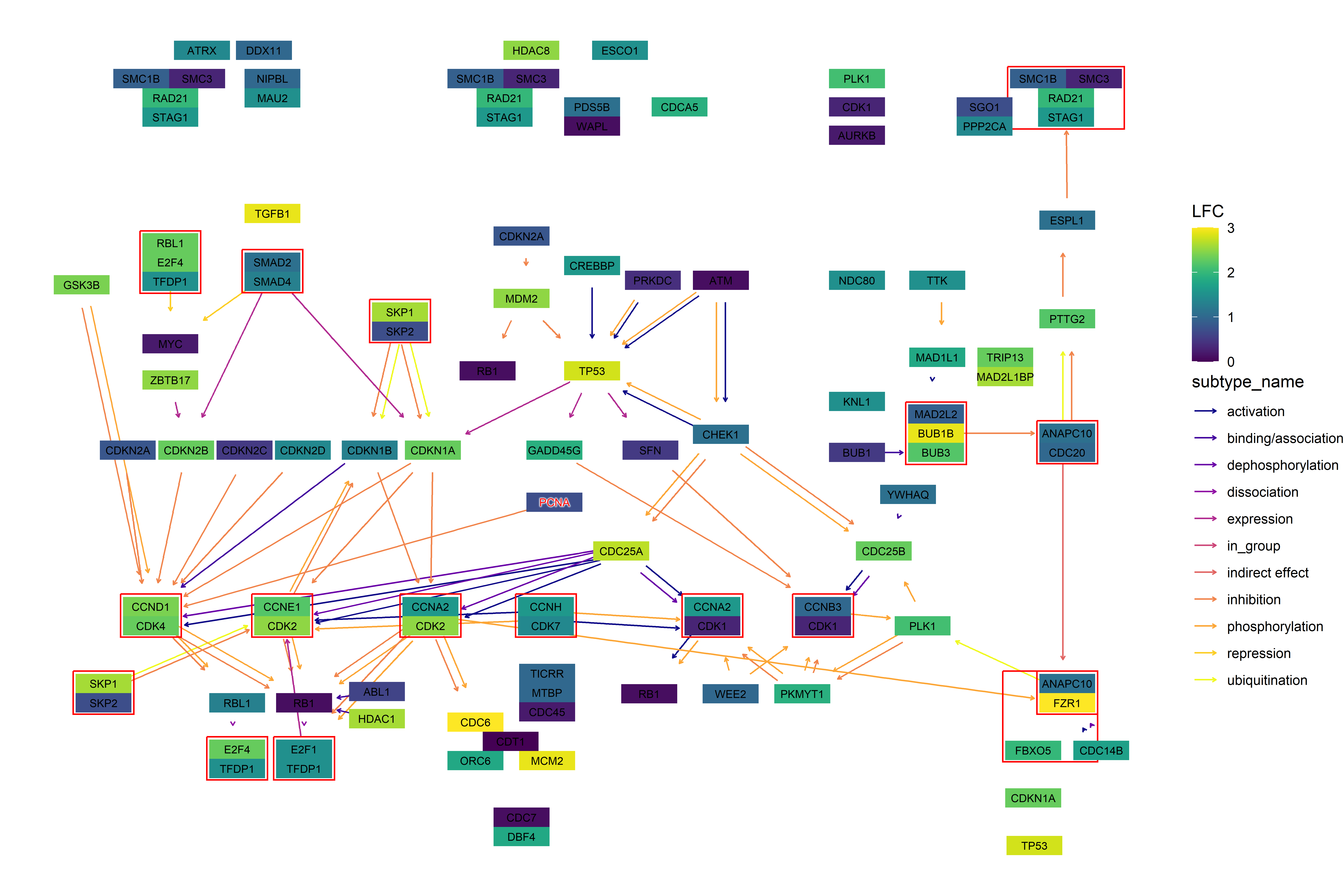
g <- pathway("hsa04110")
pseudo_lfc <- sample(seq(0,3,0.1), length(V(g)), replace=TRUE)
names(pseudo_lfc) <- V(g)$name
ggkegg("hsa04110",
convert_org = c("pathway","hsa","ko"),
numeric_attribute = pseudo_lfc)+
geom_edge_parallel2(
aes(color=subtype_name),
arrow = arrow(length = unit(1, 'mm')),
start_cap = square(1, 'cm'),
end_cap = square(1.5, 'cm')) +
geom_node_rect(aes(filter=.data$type=="group"),
fill="transparent", color="red")+
geom_node_rect(aes(fill=numeric_attribute,
filter=.data$type=="gene"))+
geom_node_text(aes(label=converted_name,
filter=.data$type == "gene"),
size=2.5,
color="black")+
with_outer_glow(geom_node_text(aes(label=converted_name,
filter=converted_name=="PCNA"),
size=2.5, color="red"),
colour="white",
expand=4)+
scale_edge_color_manual(values=viridis::plasma(11))+
scale_fill_viridis(name="LFC")+
theme_void()