This repository contains the official implementation of Cliqueformer, as described in the paper:
Cliqueformer: Model-Based Optimization with Structured Transformers
Jakub Grudzien Kuba, Pieter Abbeel, Sergey Levine
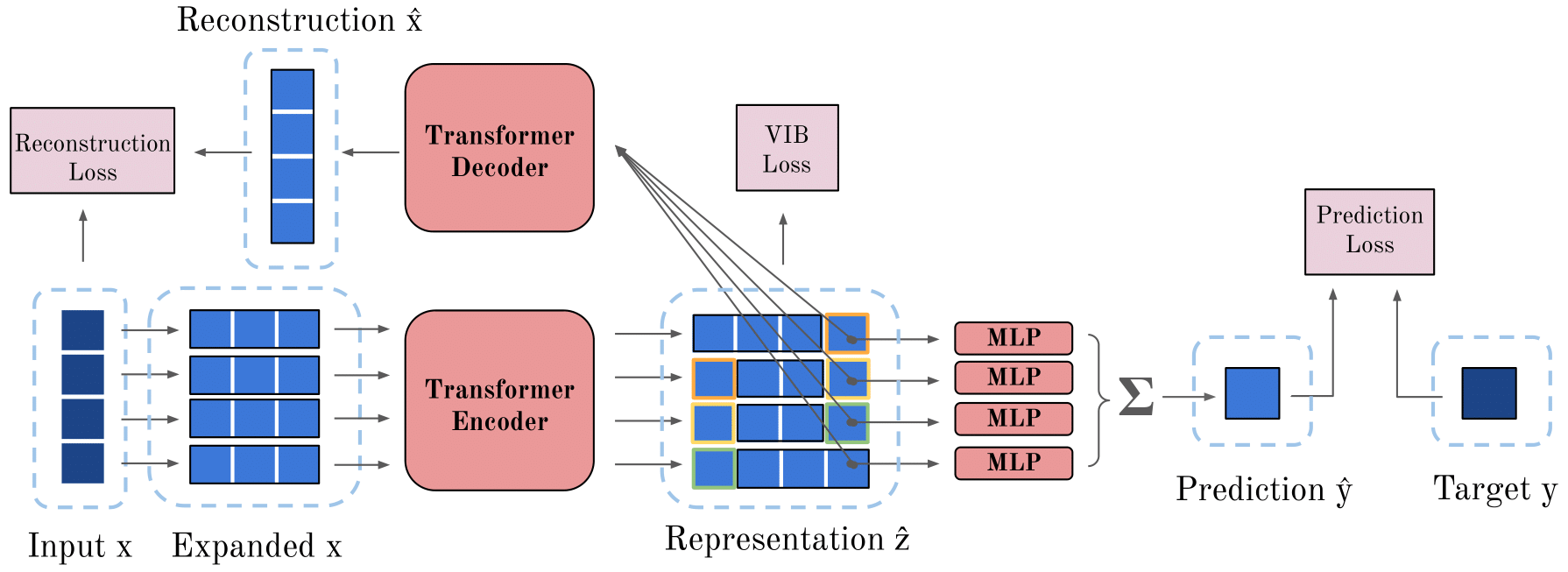
Cliqueformer is a scalable transformer-based architecture for model-based optimization (MBO) that learns the structure of the black-box function in the form of its functional graphical model (FGM). Cliqueformer demonstrates state-of-the-art performance on various tasks, from high-dimensional black-box functions to real-world chemical and genetic design problems.
- Learns the structure of MBO tasks through functional graphical models
- Scalable transformer-based architecture
- Outperforms existing methods on benchmark tasks
git clone https://github.com/znowu/cliqueformer-code.git
cd cliqueformer-code
pip install -r requirements.txtcd scrape/Bioseq
python download_data.py
python download_model.py
cd ../..python training.pypython optimize.pyYou can change the task you want to solve by changing the config file in training.py and optimize.py. For example, for Superconductor: 'configs/superconductor/cliqueformer.py'.