R package developed for statistical analysis isotops developped in the frame of the ANR-Itineris
Install the R package
devtools::install_github("zoometh/itineRis")
And load it
library(itineRis)
By default, output will be saved in the results/ folder. You can change the output folder by changing the dirOut option in the various functions.
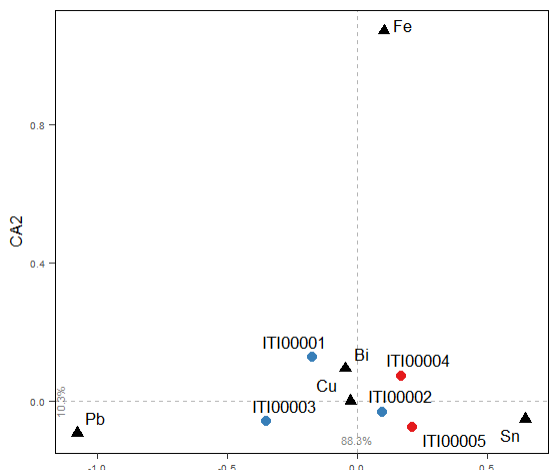
Run the isotop_ca() function to compute Correspondence Analysis (CA)
isotop_ca(col.group = "Provenienza",
pt_siz = 2,
lbl.size = 3)
The user can select 3 or more variables that appears both in the dataset and in the reference thesaurus. The latter gathers lists of elements (Cu, Ar, ...), chemical compounds (SiO2, TiO2, ...), or isotopes.
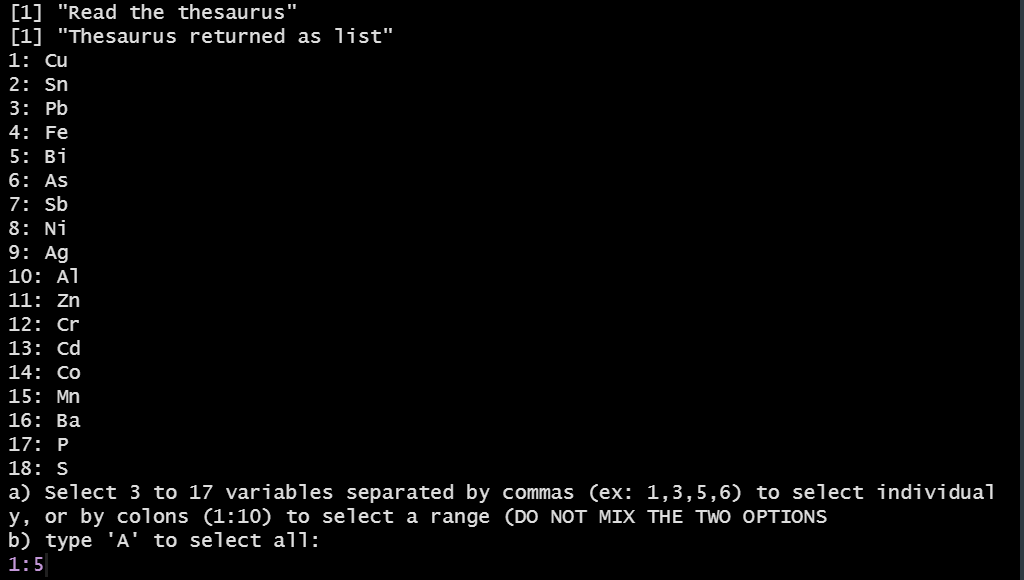
Selection of variables (elements)
The result will is:
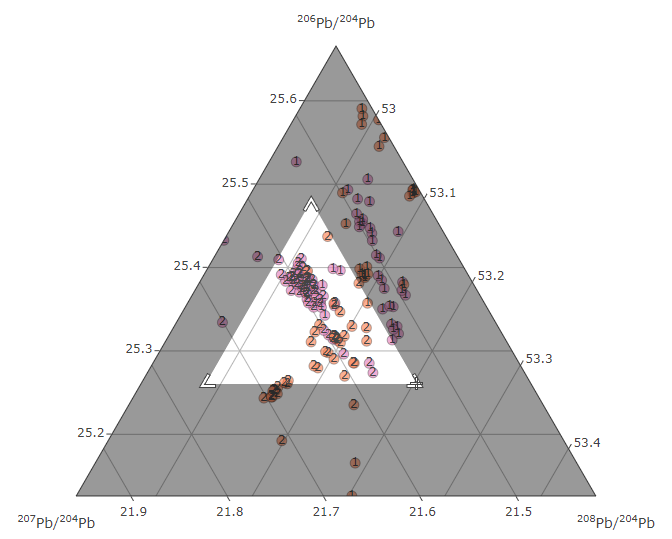
Create an interactive ternary plot, with plotly, to visualise the results of lead isotopic analysis (LIA)
Create an hash object to store
d <- hash::hash()
Read the LIA measures for objects and mines
d <- read_lia(d = d,
d.tag = "lia.objects",
df.path = "C:/Rprojects/itineRis/results/LIA data objects.xlsx",
header.line = 2)
d <- read_lia(d = d,
d.tag = "lia.mines",
pattern.objects.num = "Locality/.Mine",
pattern.objects.Pb206_Pb204 = "206Pb/204Pb",
pattern.objects.Pb207_Pb204 = "207Pb/204Pb",
pattern.objects.Pb208_Pb204 = "208Pb/204Pb",
df.path = "C:/Rprojects/itineRis/results/Coordinates-mines_Thomas Huet.xlsx",
header.line = 1)
Group the datasets, assign colors and symbols
library(dplyr)
mydf <- rbind(d$lia.mines, d$lia.objects)
df.isotop <- isotop_dataframe(df = mydf, df.path = NA)
df.isotop[df.isotop$object == "golasecca", "color.object"] <- "#0000FF"
df.isotop[df.isotop$object == "hochdorf", "color.object"] <- "#00FFFF"
df.isotop[df.isotop$object == "France", "color.object"] <- "#FF0000"
df.isotop[df.isotop$object == "France", "symbol"] <- "triangle"
df.isotop[df.isotop$object == "Iberian Peninsula", "color.object"] <- "#FFA500"
df.isotop[df.isotop$object == "Iberian Peninsula", "symbol"] <- "triangle"
df.isotop[df.isotop$object == "Switzerland", "color.object"] <- "#964B00"
df.isotop[df.isotop$object == "Switzerland", "symbol"] <- "triangle"
Create a ternary plot
isotop_ternaryplot(df.isotop)
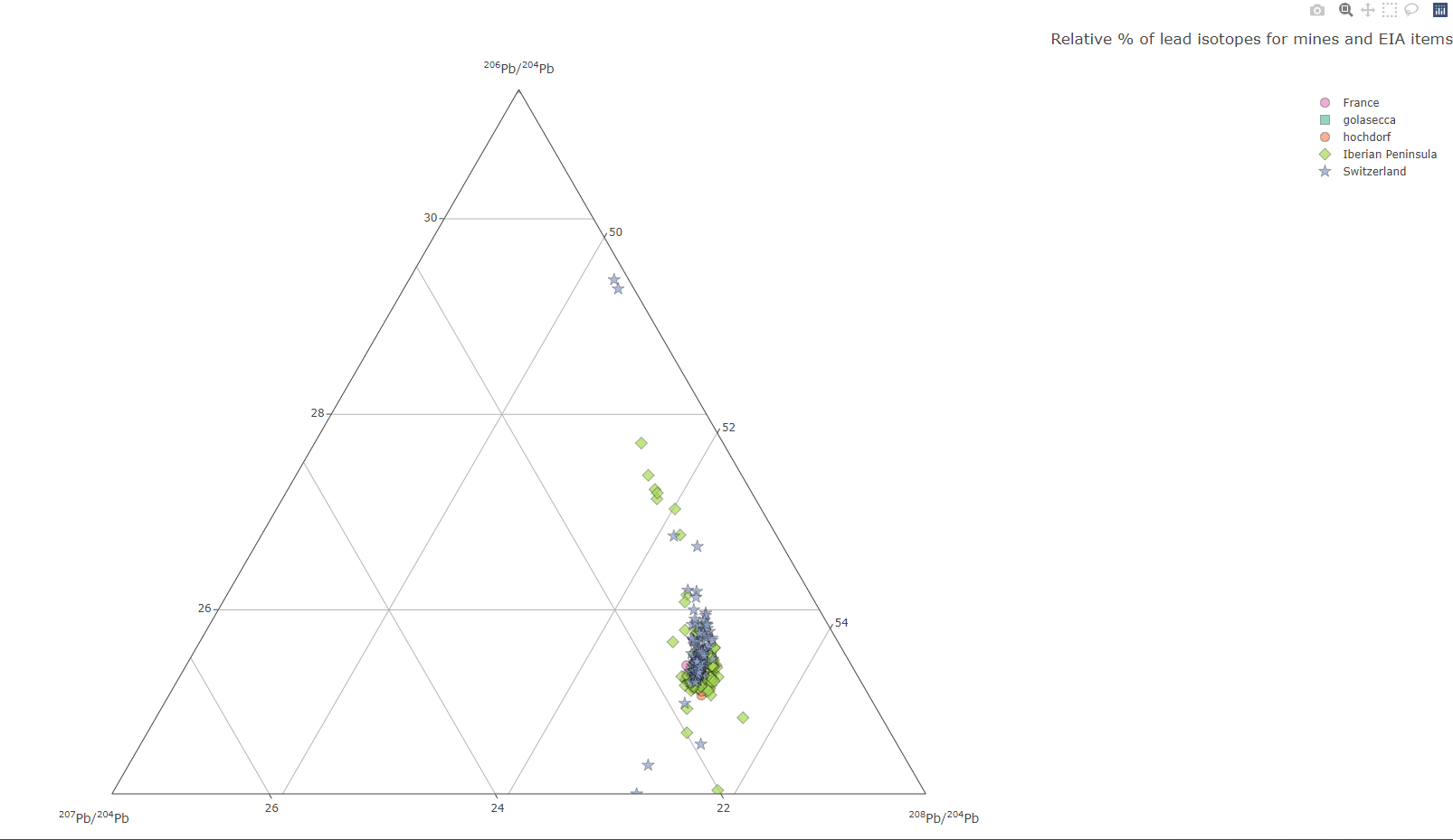
landing page of the interactive Plotly plot
A similar function is hosted on a Shiny Server: http://shinyserver.cfs.unipi.it:3838/teach/stats/stats/dim3/
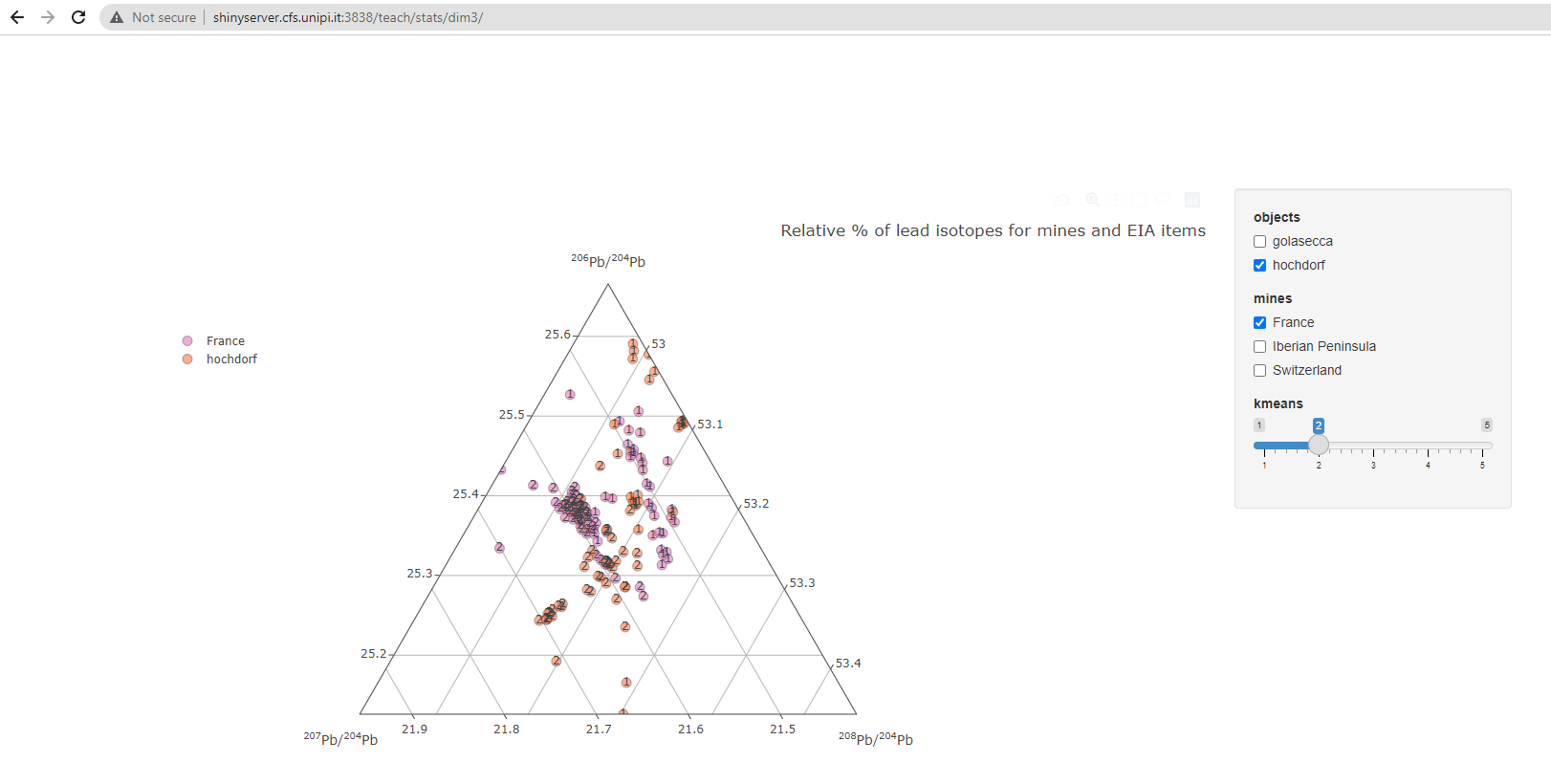
landing page of the interactive Shiny ternary plot
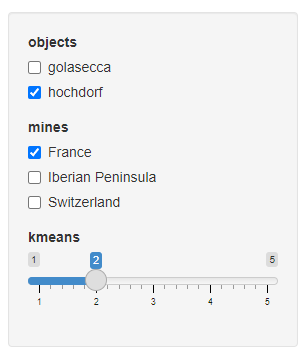
The parameters (objects, mines, kmeans) can be selected with the input box:
objects: categories of objects (by defaultgolaseccaorhordorf)mines: mines' countries (by defaultFrance,Iberian Peninsula, orSwitzerland)kmeans: number of Kmeans1 centers, or clusters (by default between1to5)
Use the Zoom tool to select a region of interest in the graph (to zoom out, double click).
The Zoom tool, the Span tool and the Hover info allow you to view the results sample by sample.
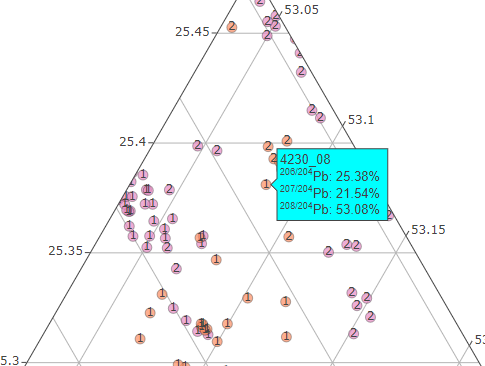
hover info for the sample `4230_08` allocated to the cluster `1`
Each sample is tagged with the number of the Kmeans cluster to which it belongs.
Create an interactive leaflet map of mines
d <- hash::hash()
d <- read_mines(d = d,
df.path = "C:/Rprojects/itineRis/results/Coordinates-mines_Thomas Huet.xlsx")
map_leaflet(d = d,
d.coords = "mines.coords",
export.plot = T,
out.plot = "map_sites.html",
dirOut = "C:/Rdev-itineris/itineris/data/")
Gives:
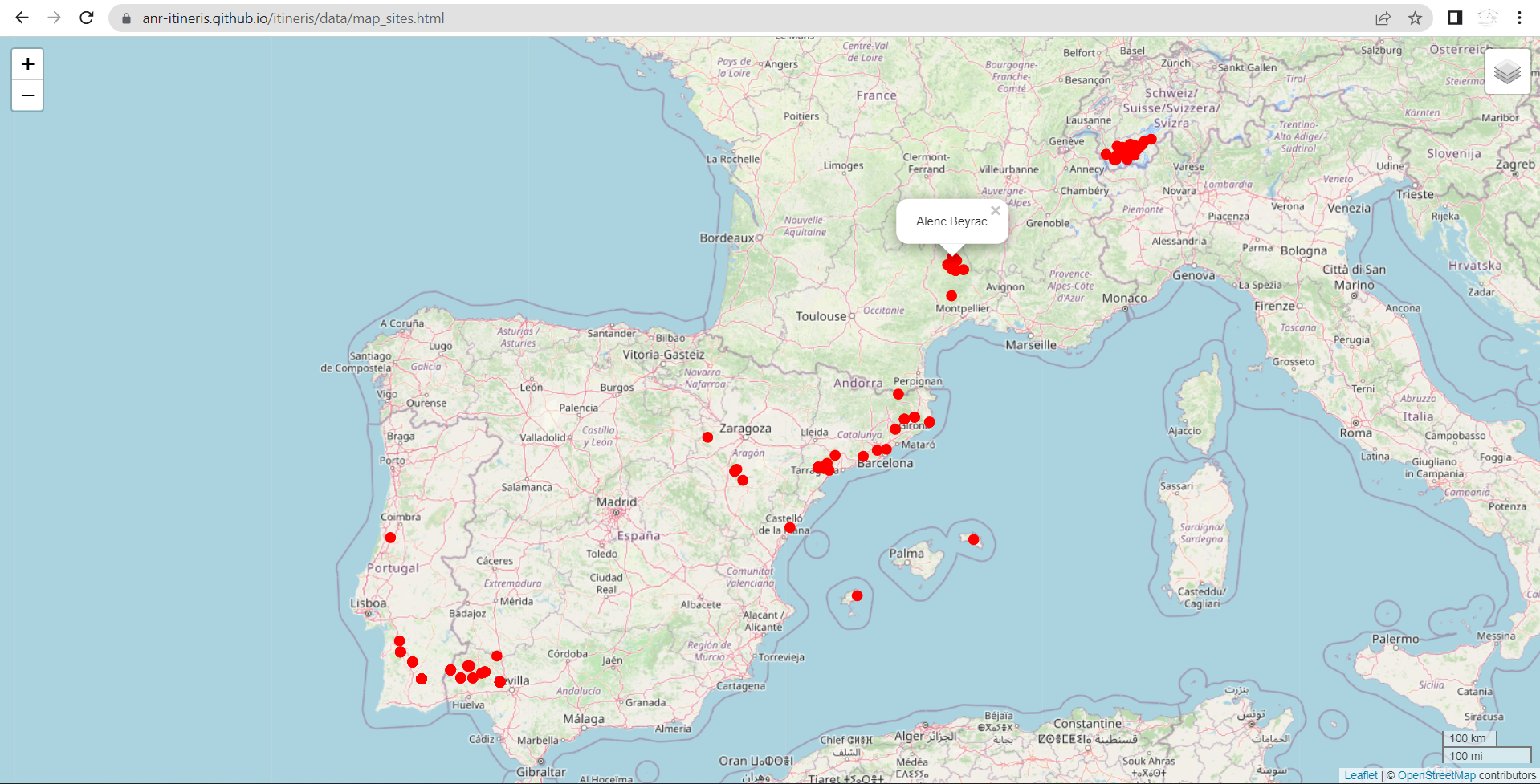
https://zoometh.github.io/itineRis/results/map_sites.html
The function thesaurus() creates interactive graphs to display the thesaurus of the project
thes <- "https://raw.githubusercontent.com/ANR-Itineris/itineris/main/lod/thesaurus/Itineris_th101.rdf"
thesaurus(root = "artefact",
outPlot = "collapsibleTree",
export.plot = T,
outDir = "C:/Rprojects/itineRis/results/")
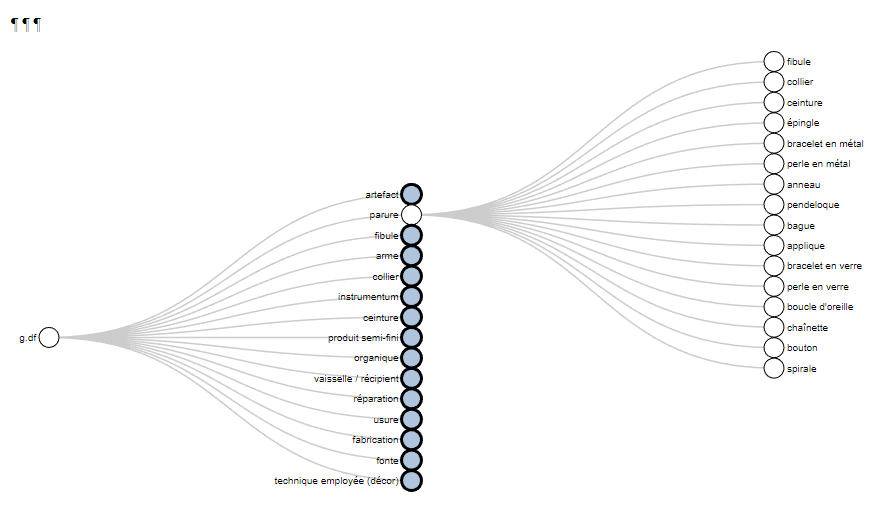
https://zoometh.github.io/itineRis/results/artefact
Footnotes
-
the Kmeans (R function
kmeans()) is run on the selectedobjectsandmines. It tags every sample with the number of its cluster () ↩