Official implementation of "Med-Unic: unifying cross-lingual medical vision-language pre-training by diminishing bias"
Source code for the paper entitled Med-UniC: Unifying Cross-Lingual Medical Vision-Language Pre-Training by Diminishing Bias
Zhongwei Wan, Che Liu, Mi Zhang, Jie Fu, Benyou Wang, Sibo Cheng, Lei Ma, César Quilodrán-Casas, Rossella Arcucci
The Ohio State University and Imperial College London, etc.
Our experiment is build on the framework from huggingface transformers.4.9.0
The main framework of our Med-UniC is shown in the figure below. 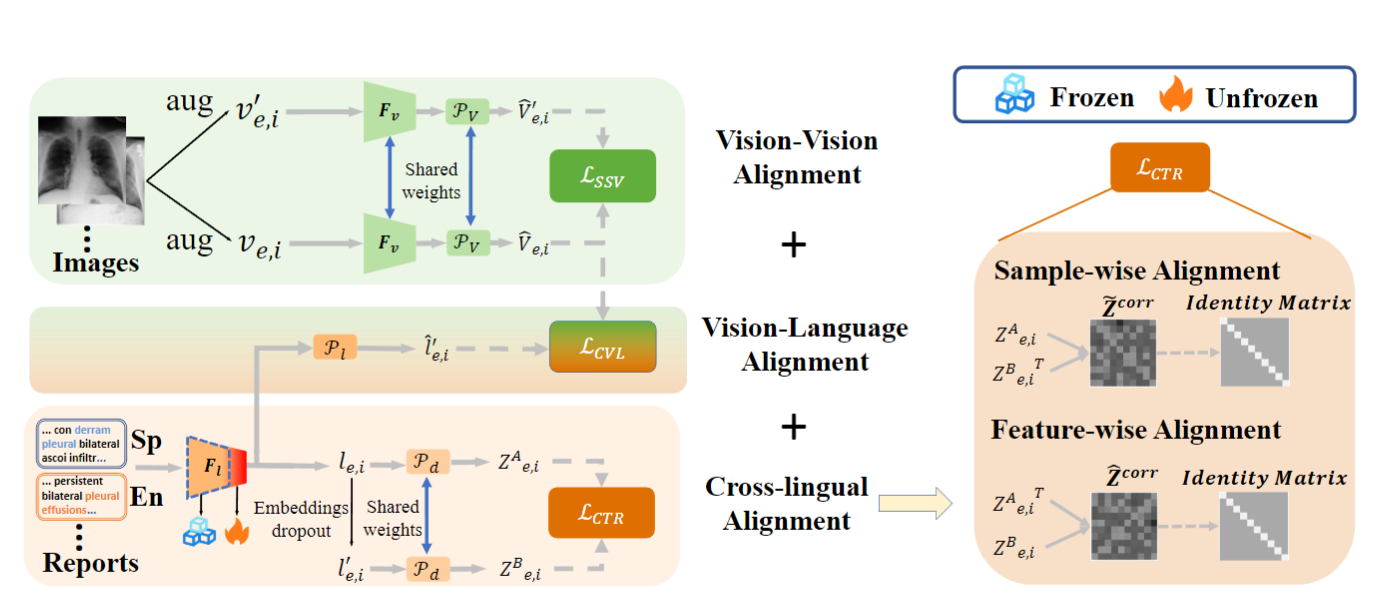
pip install -r requirement.txt
cd finetune/downstream_tasks
pip install -r requirement.txt
- Build cross-lingual vocab: Construct mixed vocab from Spanish medical corpus.
- Post-pretrain Cross-lingual Medical LM: Use MLM to post-training medical LM to acquire initial cross-lingual ability.
- Pretrain Med-UniC: Vision-language pretraining for Med-UniC.
- Downstream tasks: Finetune, zeroshot.
- Download MIMIC-CXR and PadChest datasets. For MIMIC-CXR dataset, please follow MGCA to download and obtain the 'master.csv' file. For PadChest, the data can be download from PadChest.
- To preprocess the images, please run
python preprocess.py --dataset=MIMIC(PDC) - Download checkpoint **CXR-BERT-general: **huggingface transformers. Build cross-lingual vocab:
cd generate_mix_corpus/
1.Convert csv to json:
python convert_csv_to_json_en.py
python convert_csv_to_json_sp.py
2. Generate Spanish Vocab:
python generate_CXRBert_vocab.py
3. Merge mixed vocab and replace vocab.txt of CXRBert:
python build_sp_vocab.py
4. Mix en and sp jsons for pretraining:
python mix_json.py
5. tokenize jsons for MLM:
python tokenize_pretrain_data.py
python starter_pretrain_cxrbert.py --cache_dir /cache --epochs 15 --gradient_accumulation_steps 16 --learning_rate 5e-4 load_model 1 --mask_ratio 0.15 --max_seq_length 256 --model /CXR_bert_general --nas_output_dir multiligual_cxrbert_015_15_8/ --nnodes 1 --nproc_per_node 8 --pregenerated_data tokenized_parts_med_data/ --warmup_steps 50
Arguments:
pregenerated_data: pretrained cross-lingual corpus frompython tokenize_pretrain_data.py.
Post-pretraining results:
| Hyper-params | Loss |
|---|---|
| 5 epochs 8 cards 1024 bz | 0.6938 |
| 5 epochs 16 cards 2048 bz | 0.6715 |
| 10 epochs 8 cards 1024 bz | 0.5543 |
| 10 epochs 16 cards 2048 bz | 0.5613 |
| 15 epochs 8 cards 1024 bz | 0.5296 |
| 15 epochs 16 cards 2048 bz | 0.5459 |
Then, adopt well-pretrained Cross-lingual Medical LM, run:
python starter_pretrain_mmodal.py --batch_size=128 --cache_dir=/cache --en_img_path=/nas/wanzhongwei_med_data/english_pretrain/only_imp.npy --en_text_csv_path=/nas/wanzhongwei_med_data/english_pretrain/200k_find_imp.csv --freeze_layers=9 --from_scratch=0 --gradient_accumulation_steps=4 --img_data=s3://bucket-884/wanzhongwei_multi_modal/simplified_code/img_data/ --lambda_t=1 --loss_type=unified_loss --lr=4e-5 --max_epochs=100 --max_seq_length=256 --model=/nas/wanzhongwei_med_data/cxrbert/cxrbert_15_8/ --nas_output_dir=/nas/wanzhongwei_med_data/vit_pretrain_model_128/ --nnodes=1 --nproc_per_node=8 --sp_img_path=/nas/wanzhongwei_med_data/sp_pretrain/PDC_train_int.npy --sp_text_csv_path=/nas/wanzhongwei_med_data/sp_pretrain/PDC_cleaned.csv --text_aug=0 --text_data=s3://bucket-884/wanzhongwei_multi_modal/simplified_code/new_data/ --un_pretrain_model=/nas/wanzhongwei_med_data/unpretrain_cxrbert/ --vision_encoder_name=vit --vision_model_path=/nas/wanzhongwei_med_data/well_pretrain_models/resnet50_imagnet/resnet50imageNet.pth --vit_name=base --vit_path=/nas/wanzhongwei_med_data/VIT_backbone/vit_base/pretrain_vit_base.pth --weight_decay=5e-2
Arguments:
img_data: Location to store pretraining medical sp and en image data.
Finetuning dataset downloading:
-
CheXpert: CheXpert dataset in the CheXpert .
-
RSNA: RSNA dataset in Kaggle.
-
COVIDx: COVIDx dataset in Kaggle.
-
SIIM: SIIM dataset in Kaggle.
-
Object-CXR: object-CXR dataset in its official website.
Set up the downstream tasks, preprocess the datasets, and finetune the model following MGCAhttps://github.com/HKU-MedAI/MGCA.
Then, use Med-UniC visual encoder checkpoint learned from Pretrain Med-UniC section to execute downstream tasks as following:
cd finetune/downstream_tasks/mgca/models/mgca/
CUDA_VISIBLE_DEVICES=1 python med_unic_vit_finetuner.py --gpus 1 --dataset rsna --data_pct 0.01 --batch_size 8 --seed 42
CUDA_VISIBLE_DEVICES=0 python med_unic_vit_finetuner.py --gpus 1 --dataset rsna --data_pct 0.1 --batch_size 8 --seed 42
CUDA_VISIBLE_DEVICES=0 python med_unic_vit_finetuner.py --gpus 1 --dataset rsna --data_pct 1 --batch_size 8 --seed 42
- For English Dataset zero-shot classification task, we use the dataset from the test set of CheXlocalize. It includes 500+ CXR images with clinician annotated disease label.
- For Spanish Dataset, we build the test set only with unique label. Preprocess Spanish PDC zeroshot dataset as following:
python Med-UniC/zero-shot/preprocess_pdc.py
- Zeroshot tasks:
1. CheXpert zeroshot:
python zero_shot.py
1. PDC zeroshot:
python zero_shot_pdc.py
@article{Wan2023MedUniCUC,
title={Med-UniC: Unifying Cross-Lingual Medical Vision-Language Pre-Training by Diminishing Bias},
author={Zhongwei Wan and Che Liu and Mi Zhang and Jie Fu and Benyou Wang and Sibo Cheng and Lei Ma and C'esar Quilodr'an-Casas and Rossella Arcucci},
journal={ArXiv},
year={2023},
volume={abs/2305.19894}
}