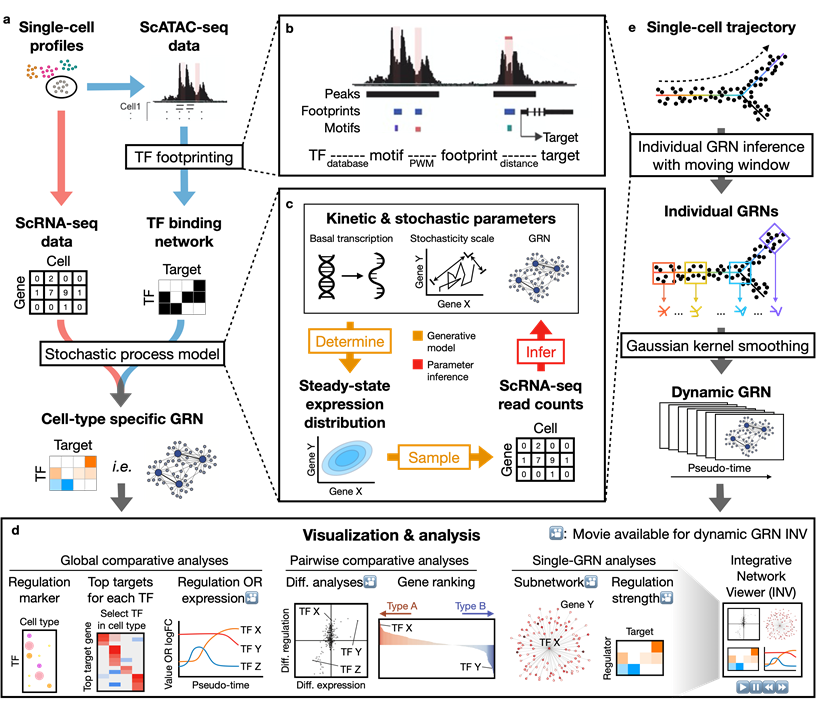
Dictys reconstructs cell-type specific and dynamic gene regulatory networks (GRN) from scRNA-seq and scATAC-seq datasets. Dictys first infers a Transcription Factor (TF) binding network with TF footprinting from single-cell chromatin accessibility. Then Dictys refines the edges with single-cell transcriptome. Dictys addresses traditional challenges in network inference by orienting causality with TF binding information, modeling transcriptional rate to reconstruct cycle-compatible networks, and using probabilistic programming to capture the scRNA-seq process.
Dictys provides network analysis and visualization at global (across all cell types), pairwise (between two cell types) and single GRN levels. Dictys directly quantifies TF regulatory activity from GRN and enables a series of analyses such as cell-type specific TF discovery as regulation markers, differential regulation analysis alongside differential expression, and TF regulatory program illustration through its subnetwork and top activation/repression targets. These GRN-based analyses can capture unique biological insights not available from mean expression.
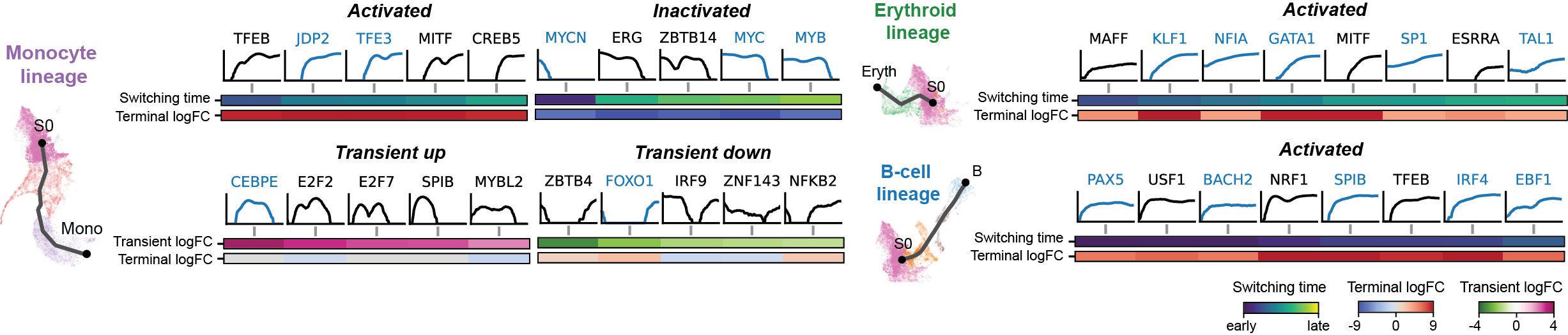
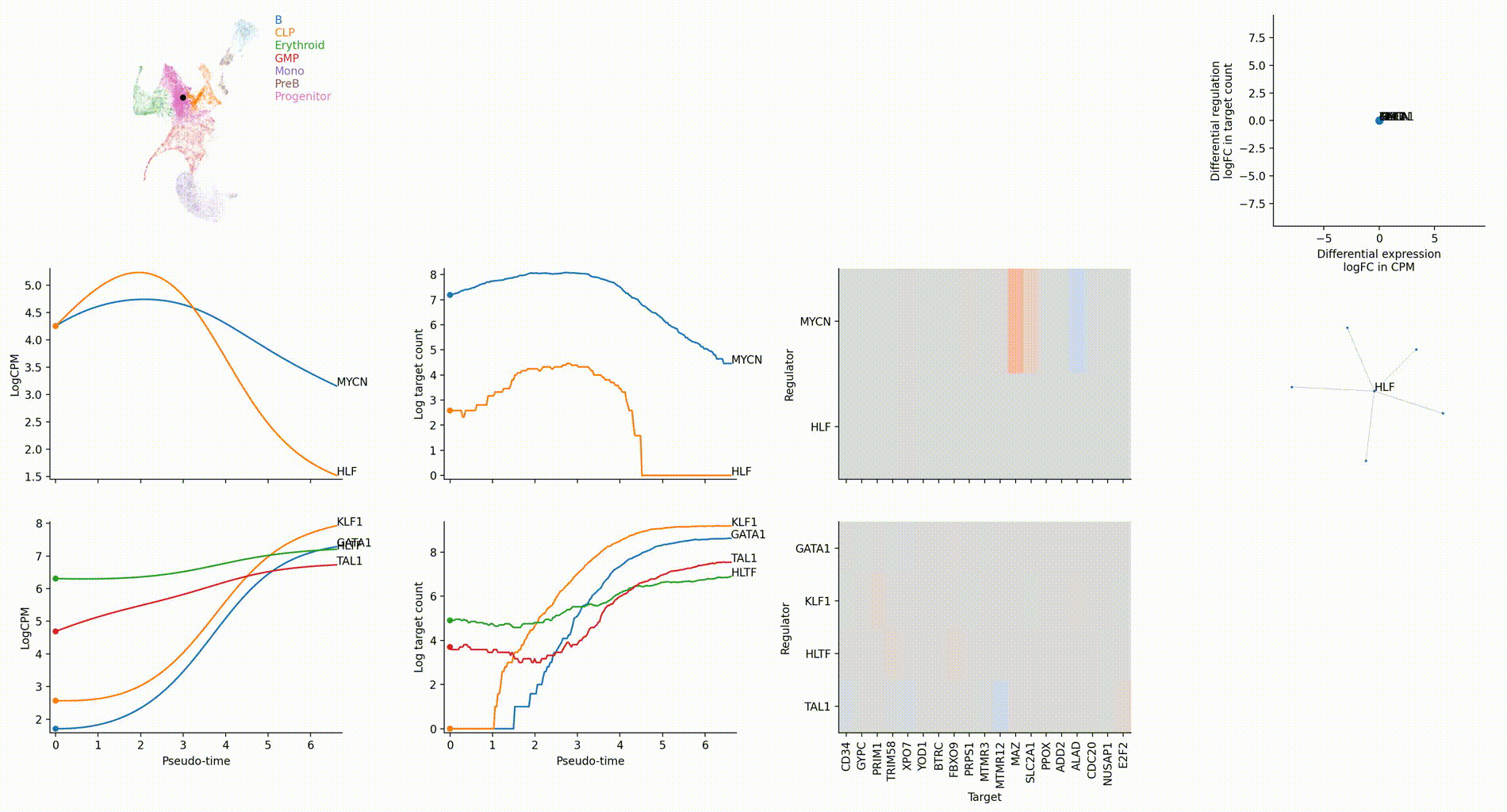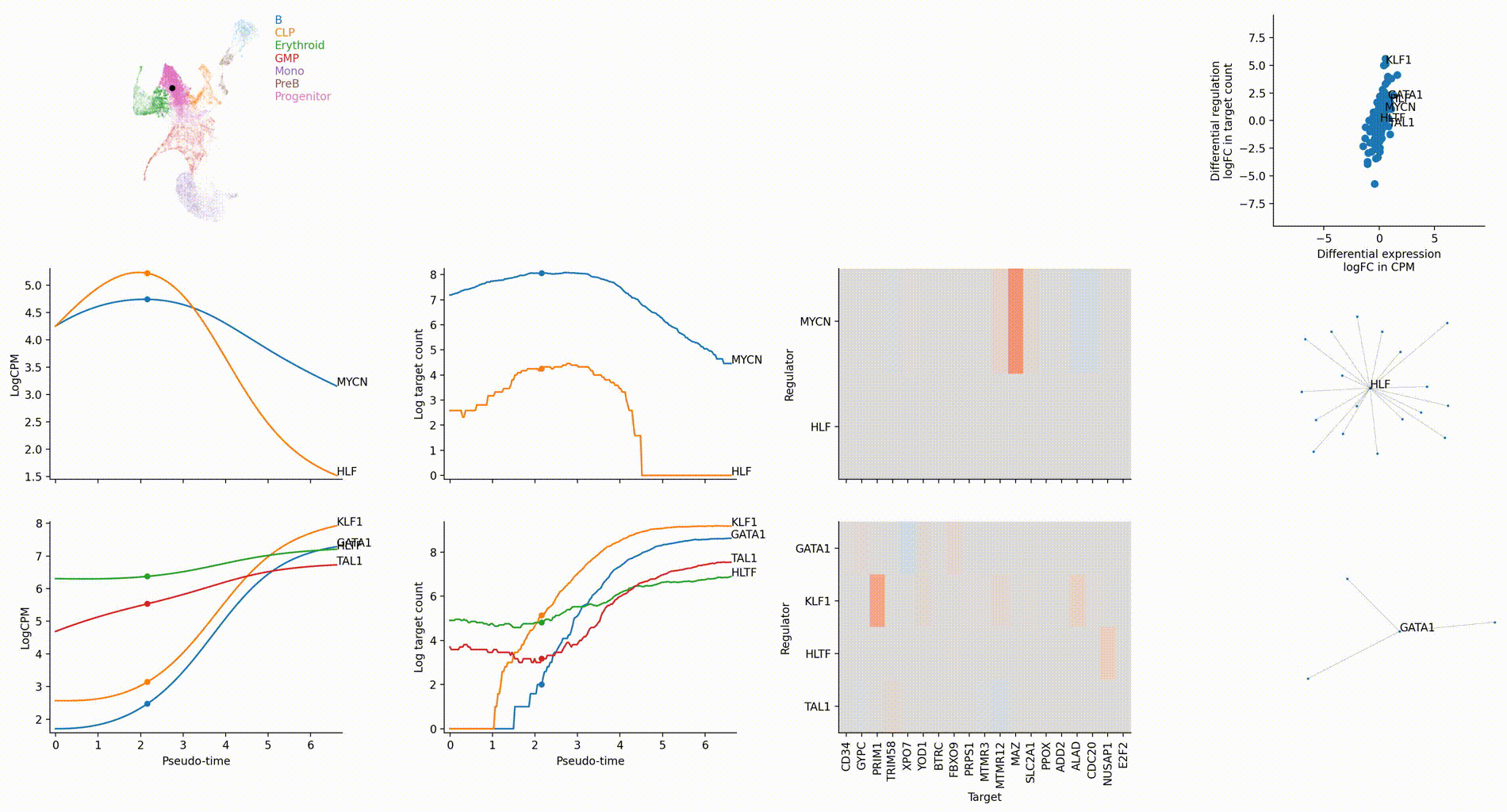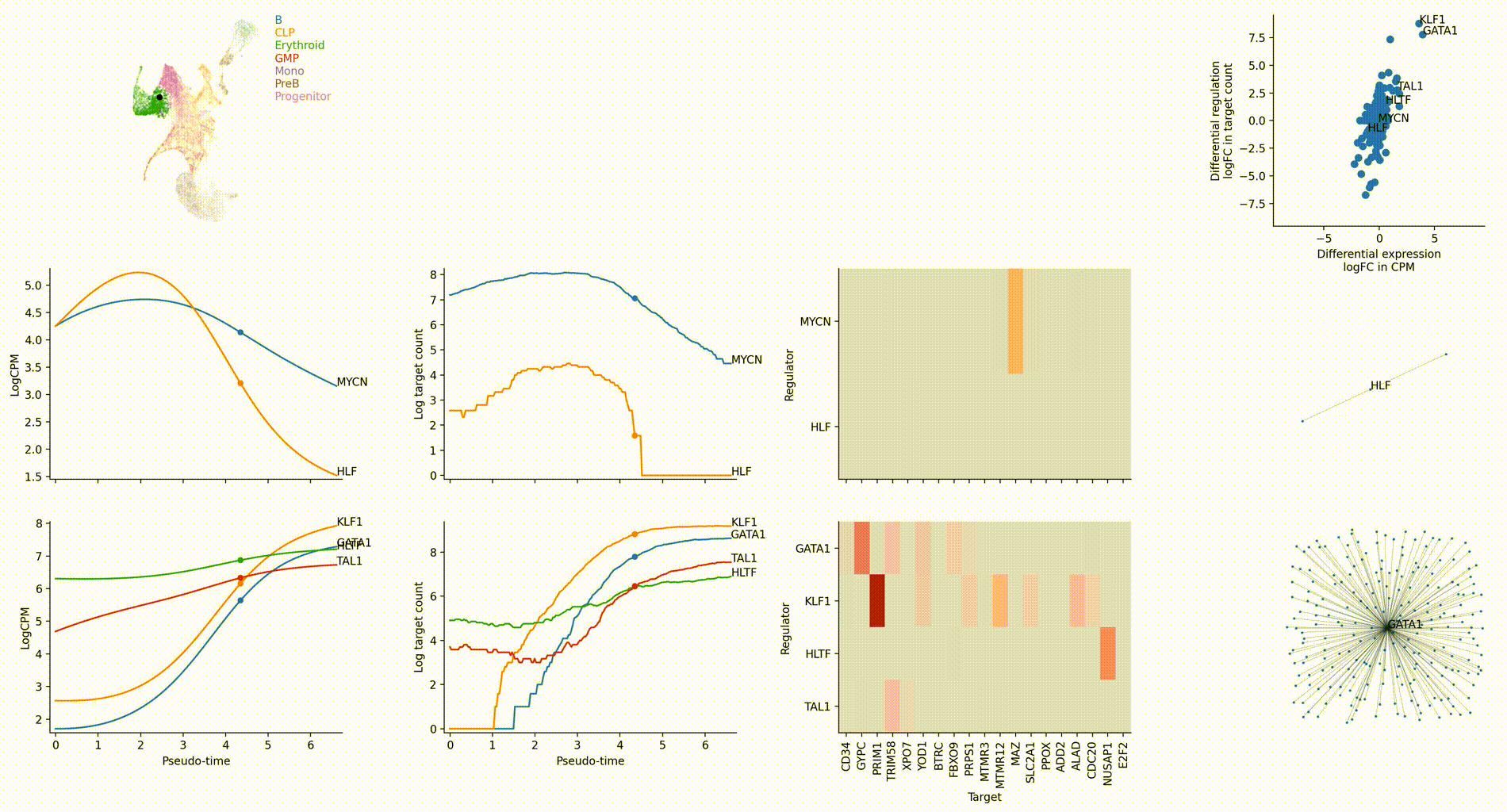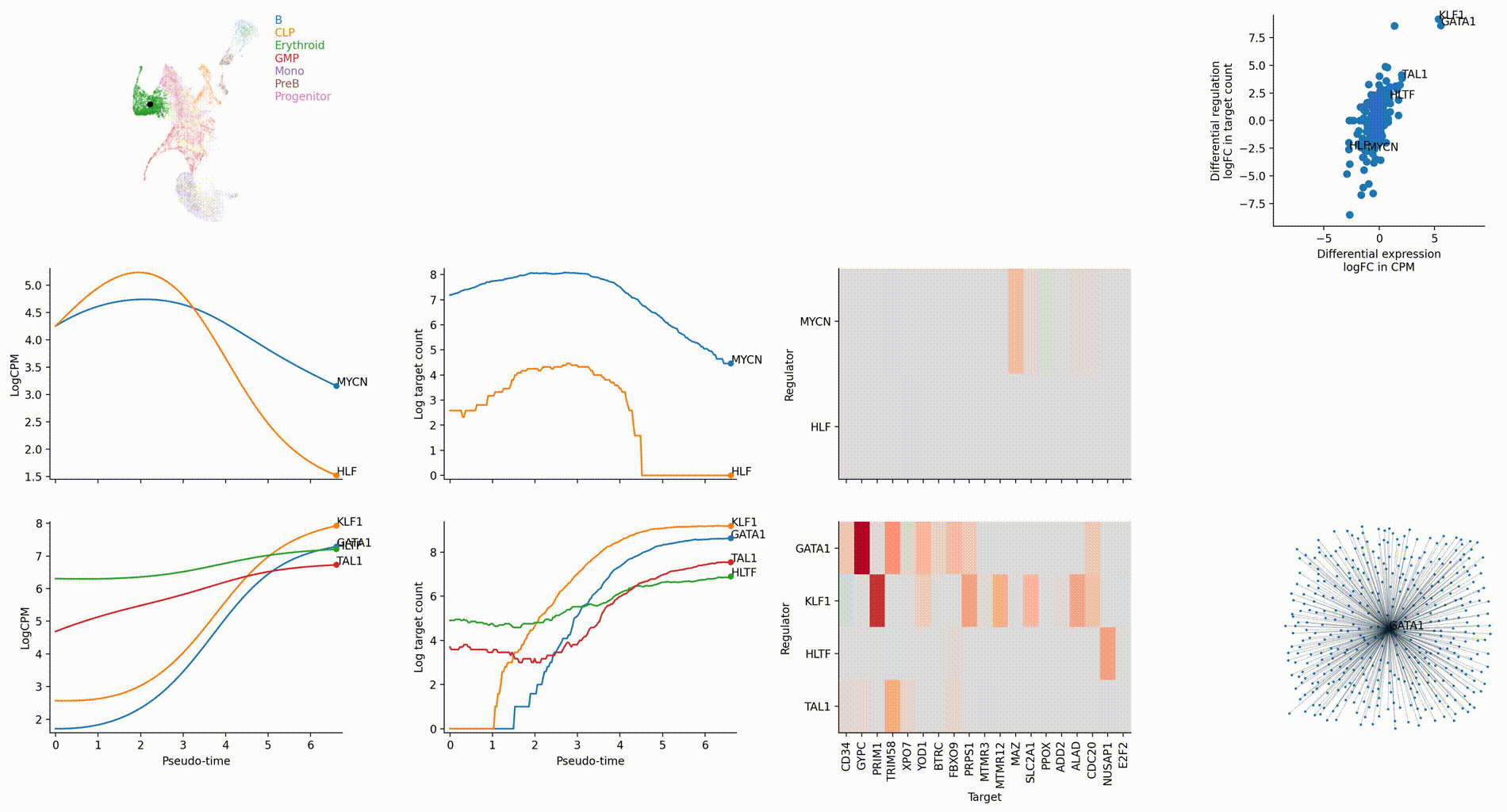
Dictys infers and analyzes dynamic GRN from scRNA-seq and scATAC-seq datasets along (inferred) trajectories. This avoids artificial cell subsets and potential biases from population imbalance, and allows (pseudo-)time-resolved discovery and investigation of driver TFs and their individual regulations. Dictys provides an integrative network viewer for dynamic GRN visualization of synchronous panels in animation.
First install Anaconda/Miniconda. Then, install Dictys with CPU computation:
conda create -y -n dictys -c conda-forge python=3.9 mamba . activate dictys mamba install -y -c lingfeiwang -c bioconda -c conda-forge -c pytorch dictys cpuonly
This will create a conda environment named dictys.
Alternatively, with GPU computation (here CUDA 11.3):
conda create -y -n dictys -c conda-forge python=3.9 mamba . activate dictys mamba install -y -c lingfeiwang -c bioconda -c conda-forge -c pytorch dictys cudatoolkit=11.3
With bash script
First install Anaconda/Miniconda. Then, install Dictys with CPU computation:
wget https://tinyurl.com/dictys -O - | bash
This will create a conda environment named dictys.
Alternatively, under a different conda environment name:
wget https://tinyurl.com/dictys -O - | CONDAENV_NAME=your_favorite_name bash
Alternatively, with GPU computation (here CUDA 11.3):
wget https://tinyurl.com/dictys -O - | CUDAVERSION_CONDA=11.3 bash
TBA
For more advanced installation, see INSTALL.md and/or edit the install script. Note: dynamic network inference is computationally intensive and GPU availability is highly recommended.
If you need STREAM, ArchR, or other softwares upstream of Dictys, we recommend to install them in separate environments following their official instructions.
We provide several tutorial for different data types. Please download each tutorial folder structure before running.
- short-multiome: a single-notebook tutorial from data preparation to context specific network analysis on 10x multiome data for human blood.
- full-multiome: an extended version of the above tutorial with detailed usage.
- analysis-blood: a simple tutorial for context specific and dynamic network analysis on separate scRNA-seq and scATAC-seq quantifications of human blood as in manuscript.
We are organizing more tutorials for release. For now, you can explore some of them without technical support on Zenodo or Google Colaboratory. Note these tutorials are subject to structural change.
The figures below are produced with the blood example dataset. You can reproduce them with the analysis-blood example. See Tutorials. Each figure is linked to the jupyter notebook that produces it.
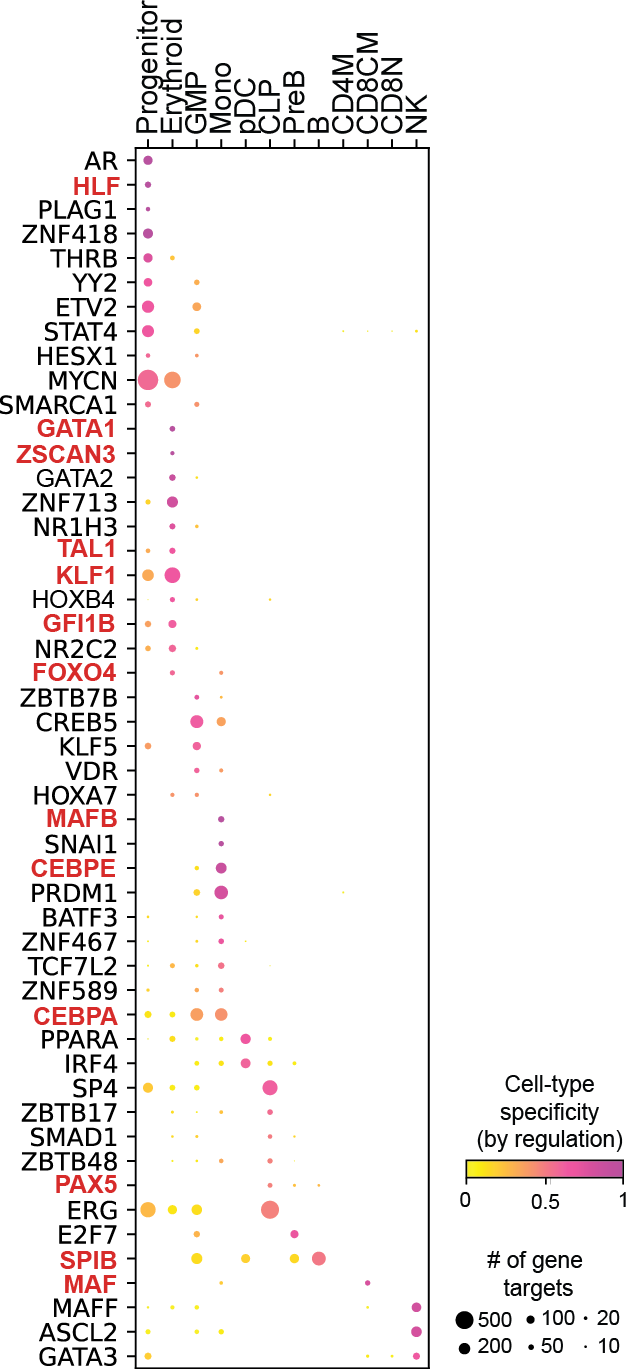
Regulation marker TF discovery
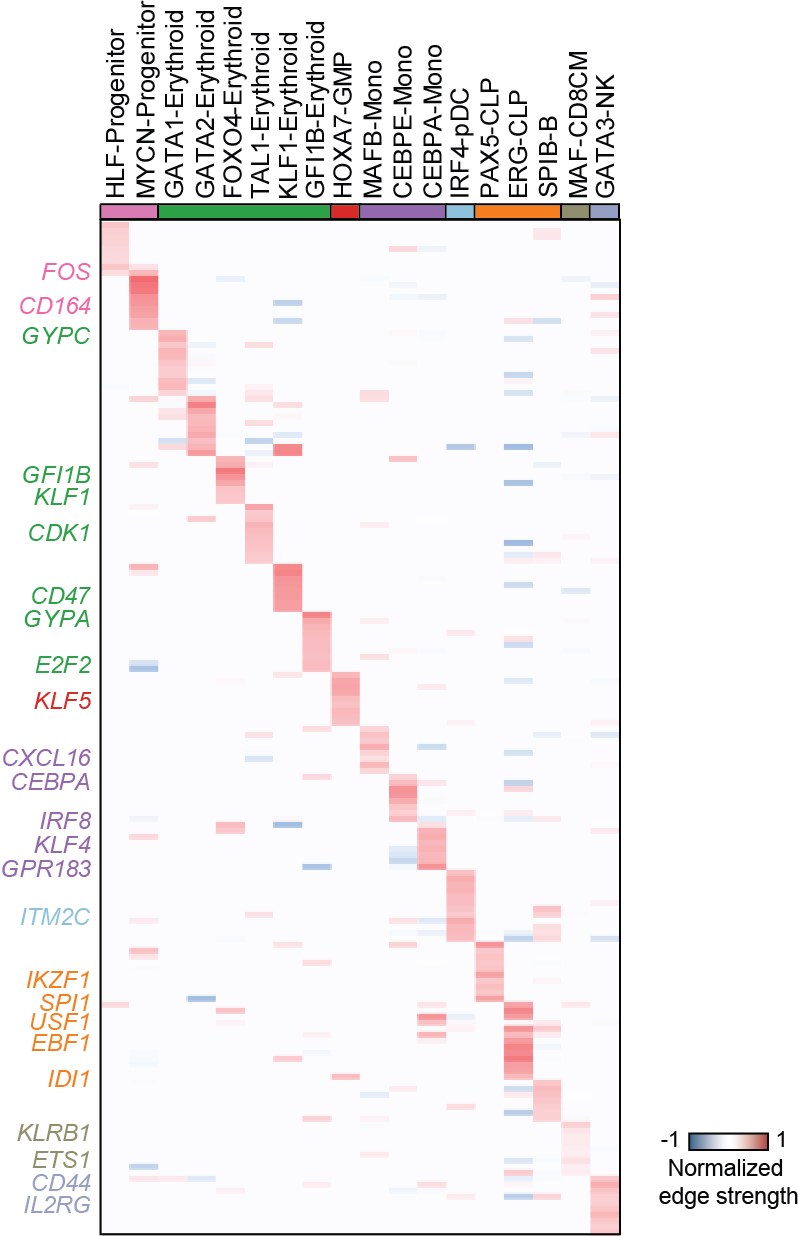
Top activation target heatmap for select TFs
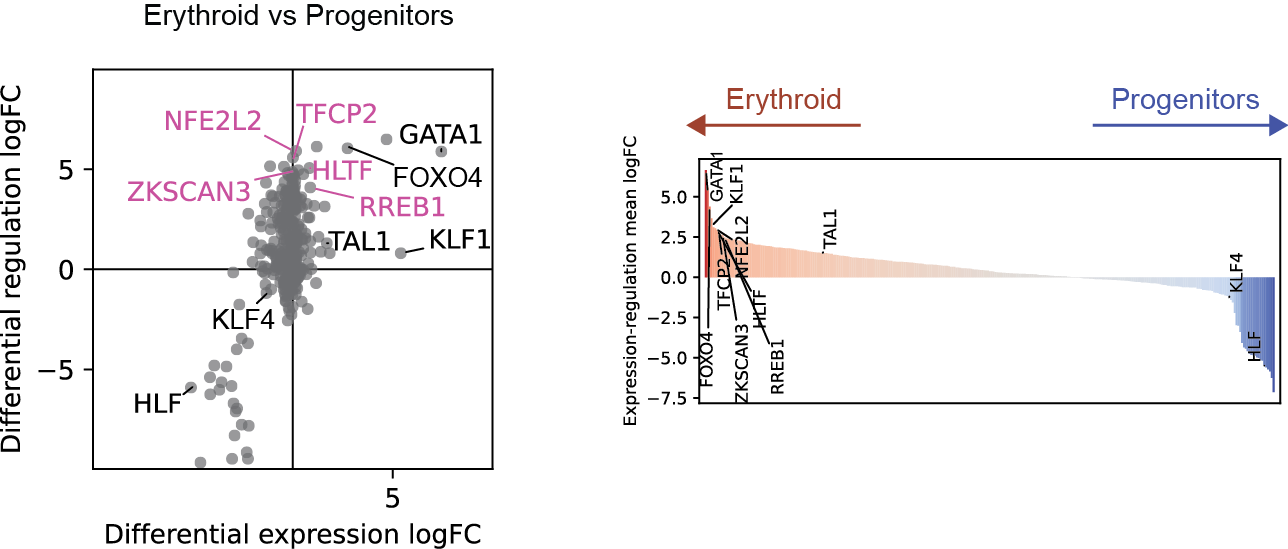
Differential regulation v.s. differential expression scatter plot; integrative TF rank plot
Driver TF discovery based on regulatory activity curve
- How do I perform network inference faster?
- Get a GPU, such as:
- Google Colaboratory offers free GPU access with zero/minimal setup. You can run Dictys on very small datasets for free, or larger datasets with paid membership. See our tutorial.
- Major cloud computing service providers offer GPU access that is orders of magnitude cheaper than a scRNA-seq experiment.
- High-performance computing cluster with GPU access at institution or other levels. Dedicated computing server. Personal computer with high-end consumer level GPU.
- People or labs with the above access.
- Reduce the computational load, such as:
- For context specific networks, choose only cell clusters of your interest. Details TBA.
- For dynamic networks, use fewer windows. This risks reducing time resolution. Details TBA.
- Reduce the number of training steps. This risks reducing network quality. Details TBA.
- Configure properly for a powerful CPU. Details TBA.
Please raise an issue on github.
Dictys: dynamic gene regulatory network dissects developmental continuum with single-cell multi-omics bioRxiv (2022)