💻 Homepage • 🤗 Dataset • 🍎 Demo • 📑 Paper • 🏖️ Overview • 🧬 Single-cell Analysis Tasks • ⌚️ QuickStart • 🛠️ Usage • 📝 Cite
ChatCell allows researchers to input instructions in either natural or single-cell language, thereby facilitating the execution of necessary tasks in single-cell analysis. Black and red texts denote human and single-cell language, respectively.- [Feb 2024] We released the model weights in small, base, and large configurations on Huggingface 🤗.
- [Feb 2024] We released the instructions of ChatCell on Huggingface 🤗.
- [Feb 2024] We released the paper.
Background
- Single-cell biology examines the intricate functions of the cells, ranging from energy production to genetic information transfer, playing a critical role in unraveling the fundamental principles of life and mechanisms influencing health and disease.
- The field has witnessed a surge in single-cell RNA sequencing (scRNA-seq) data, driven by advancements in high-throughput sequencing and reduced costs.
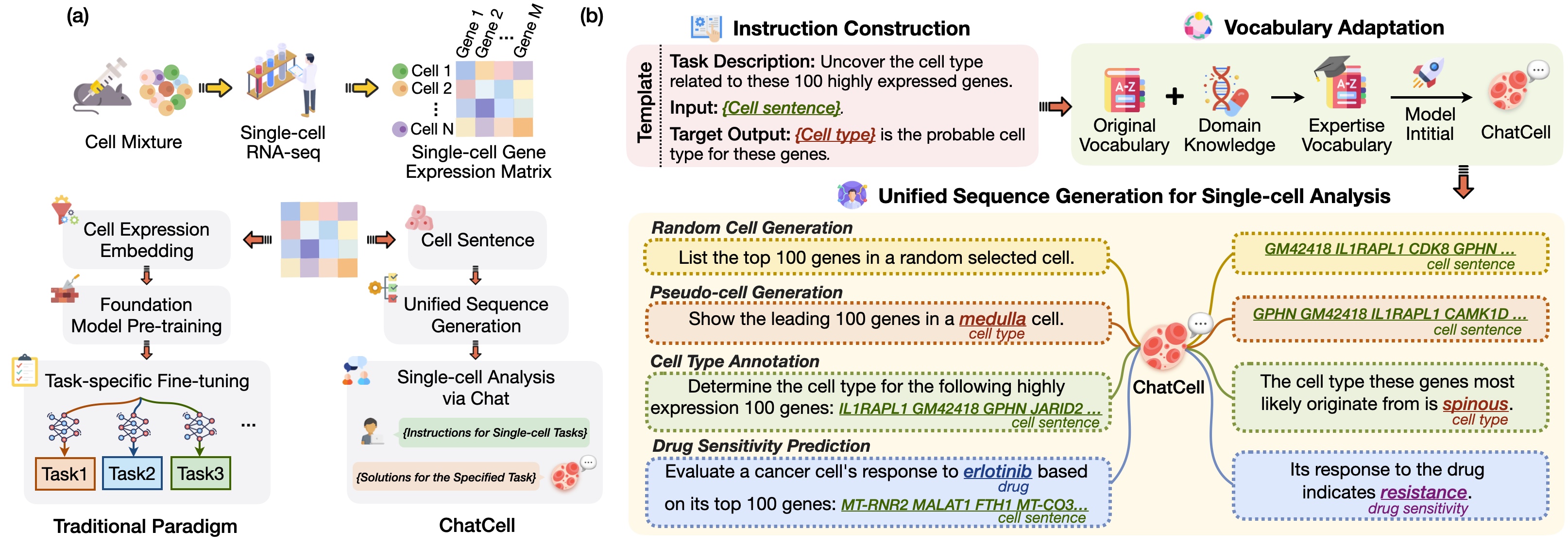
- Traditional single-cell foundation models leverage extensive scRNA-seq datasets, applying NLP techniques to analyze gene expression matrices—structured formats that simplify scRNA-seq data into computationally tractable representations—during pre-training. They are subsequently fine-tuned for distinct single-cell analysis tasks, as shown in Figure (a).
We present ChatCell, a new paradigm that leverages natural language to make single-cell analysis more accessible and intuitive.
- Initially, we convert scRNA-seq data into a single-cell language that LLMs can readily interpret.
- Subsequently, we employ templates to integrate this single-cell language with task descriptions and target outcomes, creating comprehensive single-cell instructions.
- To improve the LLM's expertise in the single-cell domain, we conduct vocabulary adaptation, enriching the model with a specialized single-cell lexicon.
- Following this, we utilize unified sequence generation to empower the model to adeptly execute a range of single-cell tasks.
We concentrate on the following single-cell tasks:
- Random Cell Sentence Generation. Random cell sentence generation challenges the model to create cell sentences devoid of predefined biological conditions or constraints. This task aims to evaluate the model's ability to generate valid and contextually appropriate cell sentences, potentially simulating natural variations in cellular behavior.
- Pseudo-cell Generation. Pseudo-cell generation focuses on generating gene sequences tailored to specific cell type labels. This task is vital for unraveling gene expression and regulation across different cell types, offering insights for medical research and disease studies, particularly in the context of diseased cell types.
- Cell Type Annotation. For cell type annotation, the model is tasked with precisely classifying cells into their respective types based on gene expression patterns encapsulated in cell sentences. This task is fundamental for understanding cellular functions and interactions within tissues and organs, playing a crucial role in developmental biology and regenerative medicine.
- Drug Sensitivity Prediction. The drug sensitivity prediction task aims to predict the response of different cells to various drugs. It is pivotal in designing effective, personalized treatment plans and contributes significantly to drug development, especially in optimizing drug efficacy and safety.
from transformers import AutoTokenizer, AutoModelForSeq2SeqLM
tokenizer = AutoTokenizer.from_pretrained("zjunlp/chatcell-small")
model = AutoModelForSeq2SeqLM.from_pretrained("zjunlp/chatcell-small")
input_text="Detail the 100 starting genes for a Mix, ranked by expression level: "
# Encode the input text and generate a response with specified generation parameters
input_ids = tokenizer(input_text,return_tensors="pt").input_ids
output_ids = model.generate(input_ids, max_length=512, num_return_sequences=1, no_repeat_ngram_size=2, top_k=50, top_p=0.95, do_sample=True)
# Decode and print the generated output text
output_text = tokenizer.decode(output_ids[0],skip_special_tokens=True)
print(output_text)❗️Note: You can download the original data from the raw_data directory. Alternatively, you can directly download the data we provide on huggingface to skip Step 1 of the process.
1. For tasks such as random cell sentence generation, pseudo-cell generation, and cell type annotation, we utilize cells from the SHARE-seq mouse skin dataset.
Follow these steps to use the transform.py script in workflow_data folder to translate scENA-seq data into cell sentence:
- Define
data_filepathto specify the path to your downloaded SHARE-seq mouse skin dataset.h5adfile. - Define
output_dirto specify directory where the generated cell sentences will be saved. - Define
eval_output_dirto specify where figures and evaluation metrics will be stored. - Run the transformation process by executing the following command in your terminal:
python transform.py.
Then covert cell sentences to instructions with mouse_to_json.py in workflow_data fold:
- Set
input_pathto theoutput_dirspecified intransform.py. - Define
train_json_file_path,val_json_file_path, andtest_json_file_pathto specify the paths where you want to save your train, validation, and test datasets in JSON format, respectively. - Run the following command in your terminal to start the conversion process:
python mouse_to_json.py.
2. For the drug sensitivity prediction task, we select GSE149383 and GSE117872 datasets.
- For GSE149383: Open
GSE149383_to_json.py, defineexpression_data_pathandcell_info_pathto the location of your downloadederl_total_data_2K.csvanderl_total_2K_meta.csvfile. - For GSE117872: Open
GSE117872_to_json.py, defineexpression_data_pathandcell_info_pathto the location of your downloadedGSE117872_good_Data_TPM.txtandGSE117872_good_Data_cellinfo.txtfile. - Update
output_json_pathwith the desired location for the JSON output files. - Execute the conversion script:
- Run
python GSE149383_to_json.pyfor the GSE149383 dataset. - Run
python GSE117872_to_json.pyfor the GSE117872 dataset.
- Run
- Open
split.py, defineinput_pathto the same locations asoutput_json_pathused above. Specify the locations fortrain_json_file_path,val_json_file_path, andtest_json_file_pathwhere you want the split datasets to be saved. - Run the script with
python split.pyto split the dataset into training, validation, and test sets.
3. After preparing instructions for each specific task, follow the steps below to merge the datasets using the merge.py script.
- Ensure that the paths for
train_json_file_path,val_json_file_path, andtest_json_file_pathare correctly set to point to the JSON files you previously generated for each dataset, such asGSE117872,GSE149383, andmouse. - Run
python merge.pyto start the merging process. This will combine the specified training, validation, and testing datasets into a unified format, ready for further analysis or model training.
1. Training
- Open the
finetune.pyscript. Update the script with the paths for your training and validation JSON files (train_json_pathandvalid_json_path), the tokenizer location (tokenizer_path), the base model directory (model_path), and the directory where you want to save the fine-tuned model (output_dir). - Execute the fine-tuning process by running the following command in your terminal:
python finetune.py
2. Generation
- Single-Instance Inference:
- To run inference on a single instance, set the necessary parameters in
inference_one.py. - Execute the script with:
python inference_one.py.
- To run inference on a single instance, set the necessary parameters in
- Web Interface Inference:
- For interactive web interface inference using Gradio, configure
inference_web.pywith the required parameters. - Launch the web demo by running:
python inference_web.py.
- For interactive web interface inference using Gradio, configure
- Batch Inference:
- For inference on a batch of instances, adjust the parameters in
inference_batch.pyas needed. - Start the batch inference process with:
python inference_batch.py.
- For inference on a batch of instances, adjust the parameters in
For the pseudo-cell generation task, we also translate sentences into gene expressions, including data extraction and transformation stages.
-
Data Extraction:
- Open
extract_gene_generation.py. Set up the necessary parameters for generating cells based on cell type. This step is intended for training datasets larger than 500 samples, covering 16 cell types. - Run the following command in your terminal to start the data extraction process:
python extract_gene_generation.py.
- Open
-
Transformation Process:
- After generating the necessary files, proceed by configuring
sentence_to_expression.pywith the appropriate parameters for the translation process. - Execute the transformation script with the command:
python sentence_to_expression.py.
- After generating the necessary files, proceed by configuring
If you use our repository, please cite the following related paper:
@article{fang2024chatcell,
title={ChatCell: Facilitating Single-Cell Analysis with Natural Language},
author={Fang, Yin and Liu, Kangwei and Zhang, Ningyu and Deng, Xinle and Yang, Penghui and Chen, Zhuo and Tang, Xiangru and Gerstein, Mark and Fan, Xiaohui and Chen, Huajun},
journal={arXiv preprint arXiv:2402.08303},
year={2024},
}