2024-2We've released ChatCell, a new paradigm that leverages natural language to make single-cell analysis more accessible and intuitive. Please visit our homepage and Github page for more information.2024-1Our paper Domain-Agnostic Molecular Generation with Chemical Feedback is accepted by ICLR 2024.2024-1Our paper Mol-Instructions: A Large-Scale Biomolecular Instruction Dataset for Large Language Models is accepted by ICLR 2024.2023-10We open-source MolGen-7b, which now supports de novo molecule generation!2023-6We open-source KnowLM, a knowledgeable LLM framework with pre-training and instruction fine-tuning code (supports multi-machine multi-GPU setup).2023-6We release Mol-Instructions, a large-scale biomolecule instruction dataset for large language models.2023-5We propose Knowledge graph-enhanced molecular contrAstive learning with fuNctional prOmpt (KANO) onNature Machine Intelligence, exploiting fundamental domain knowledge in both pre-training and fine-tuning.2023-4We provide a NLP for science paper-list at https://github.com/zjunlp/NLP4Science_Papers.2023-3We release our pre-trained and fine-tuned model on 🤗 Hugging Face at MolGen-large and MolGen-large-opt.2023-2We provide a demo on 🤗 Hugging Face at Space.
To run the codes, You can configure dependencies by restoring our environment:
conda env create -f MolGen/environment.yml -n $Your_env_name$
and then:
conda activate $Your_env_name$
You can download the pre-trained and fine-tuned models via Huggingface: MolGen-large and MolGen-large-opt.
Moreover, the dataset used for downstream tasks can be found here.
The expected structure of files is:
moldata
├── checkpoint
│ ├── molgen.pkl # pre-trained model
│ ├── syn_qed_model.pkl # fine-tuned model for QED optimization on synthetic data
│ ├── syn_plogp_model.pkl # fine-tuned model for p-logP optimization on synthetic data
│ ├── np_qed_model.pkl # fine-tuned model for QED optimization on natural product data
│ ├── np_plogp_model.pkl # fine-tuned model for p-logP optimization on natural product data
├── finetune
│ ├── np_test.csv # nature product test data
│ ├── np_train.csv # nature product train data
│ ├── plogp_test.csv # synthetic test data for plogp optimization
│ ├── qed_test.csv # synthetic test data for plogp optimization
│ └── zinc250k.csv # synthetic train data
├── generate # generate molecules
├── output # molecule candidates
└── vocab_list
└── zinc.npy # SELFIES alphabet
-
- First, preprocess the finetuning dataset by generating candidate molecules using our pre-trained model. The preprocessed data will be stored in the folder
output.
cd MolGen bash preprocess.sh- Then utilize the self-feedback paradigm. The fine-tuned model will be stored in the folder
checkpoint.
bash finetune.sh
- First, preprocess the finetuning dataset by generating candidate molecules using our pre-trained model. The preprocessed data will be stored in the folder
-
To generate molecules, run this script. Please specify the
checkpoint_pathto determine whether to use the pre-trained model or the fine-tuned model.cd MolGen bash generate.sh
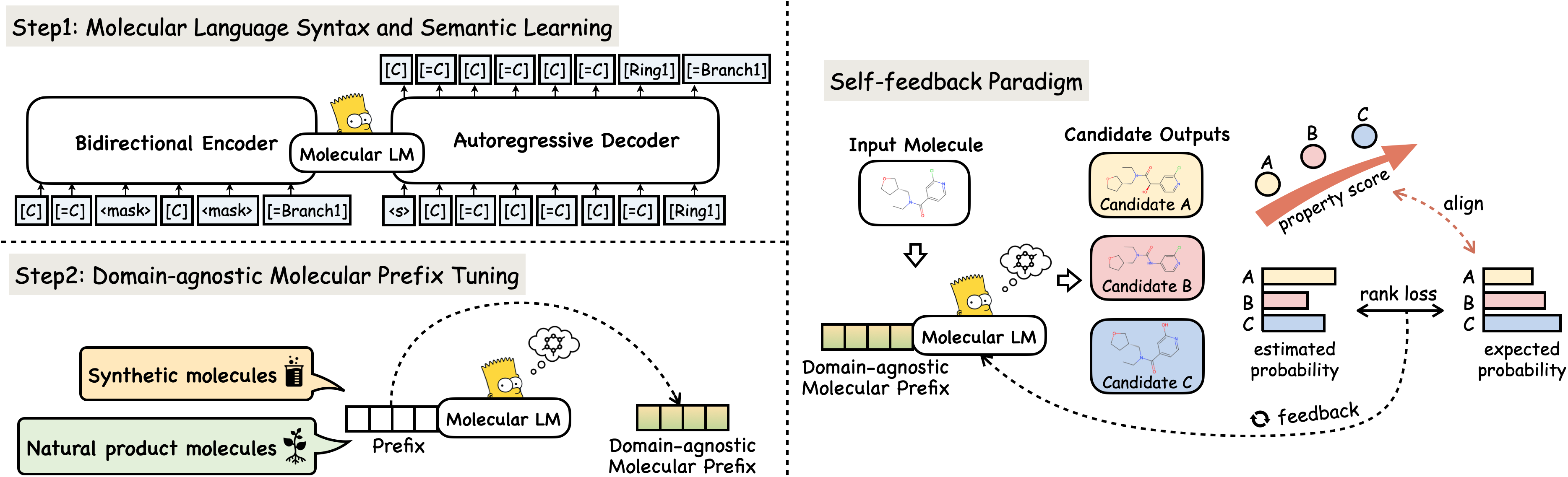
We conduct experiments on well-known benchmarks to confirm MolGen's optimization capabilities, encompassing penalized logP, QED, and molecular docking properties. For detailed experimental settings and analysis, please refer to our paper.





If you use or extend our work, please cite the paper as follows:
@inproceedings{fang2023domain,
author = {Yin Fang and
Ningyu Zhang and
Zhuo Chen and
Xiaohui Fan and
Huajun Chen},
title = {Domain-Agnostic Molecular Generation with Chemical feedback},
booktitle = {{ICLR}},
publisher = {OpenReview.net},
year = {2024},
url = {https://openreview.net/pdf?id=9rPyHyjfwP}
}