This repository contains open-source analysis approaches for the Osteoarthritis Initiative (OAI) magnetic resonance image (MRI) data. The analysis code is largely written in Python with the help of ITK and VTK for data I/O and mesh processing as well as PyTorch for the deep learning approaches for segmentation and registration. The intial development of this work was led by UNC Chapel Hill as well as Kitware. This work is also an outgrowth of conversations within the Community for Open and Reproducible Musculoskeletal Imaging Research (JC|MSK). Going forward, contributions by the broader community are, of course, not only welcome but encouraged.
The following functionality is currently supported:
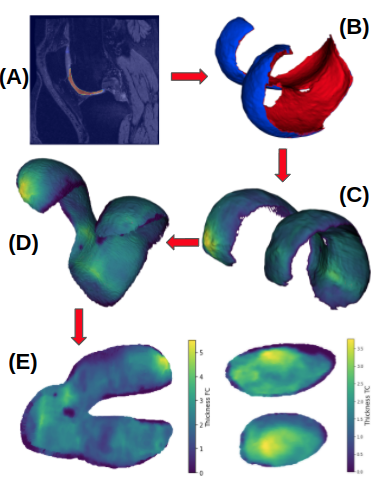
- Deep Learning Segmentation: Automatic cartilage segmentation (femoral and tibial cartilage) using a 3D UNet.
- Cartilage thickness: Extraction of femoral and tibial cartilage meshes and measuring of cartilage thickness based on a closest-point thickness estimation.
- Deep Learning Atlas registration: Registration of the carilage meshes with associated cartilage thickness to a knee atlas space via a deep registration network.
- 2D thickness mapping: Mapping of the thickness maps to a common 2D atlas space which provides full spatial correspondence. This is achieved via unrolling (based on a cylindrical coordinate system) for the femoral cartilage and a planar projection for the tibial cartilage.
- Statistical analysis: [Longitudinal statistical analysis approaches will be added shortly]
The analysis approaches in this repository are based on our initial OAI Analysis work. Much of the functionality of the original code-base has been ported to the OAI Analysis 2 repository. The main differences are
- Refactoring: A significant refactoring of the code so that it makes better use of ITK conventions as well as VTK for all the mesh processing needs.
- ICON registration: We switched to the new ICON registration approach (see manuscripts below).
- Data processing: We improved the data processing by better handling of data objects. Whereas the previous OAI Analysis pipeline largely depended on reading and writing various different files, the OAI Aanalysis 2 refactoring uses ITK and VTK objects.
- Jupyter notebooks: Better support of analysis in Jupyter notebooks.
We are currently working on the following features which should be available in the near future:
- Distributed processing: Whereas OAI Analysis was set up for cluster computing via a simple SLURM script OAI Analysis 2 is moving toward using Dask to allow for parallel processing on clusters and the cloud.
- Workflow management: Whereas OAI Analysis used custom code to avoid recomputing results, we are switching to Dagster to manage data dependencies in OAI Analysis 2.
- Distribution of analysis results: As we are planning on not only distributing code, but also analysis results (such as segmentations, meshes, thickness maps) we are planning on supporting data access via the Interplanetary File System (IPFS).
git clone https://github.com/uncbiag/oai_analysis_2
cd oai_analysis_2
pip install -r requirements.txt
python -m unittest -v discover
Currently, this should declare that the segmentation test passed, and the registration test failed.
To view the demo notebooks:
cd notebooks
jupyter notebook
upload test data to https://data.kitware.com/#collection/6205586c4acac99f42957ac3/folder/620559344acac99f42957d63
While we used the following stationary velocity field registration approach available in easyReg for OAI Analysis [paper]
@InProceedings{Shen_2019_CVPR,
title={Networks for joint affine and non-parametric image registration},
author={Shen, Zhengyang and Han, Xu and Xu, Zhenlin and Niethammer, Marc},
booktitle={Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition},
pages={4224--4233},
year={2019}
}
we now support the ICON registration approach which provides a very simple registration interface and makes use of ITK images and transforms: [paper]
@InProceedings{Greer_2021_ICCV,
author = {Greer, Hastings and Kwitt, Roland and Vialard, Francois-Xavier and Niethammer, Marc},
title = {ICON: Learning Regular Maps Through Inverse Consistency},
booktitle = {Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV)},
month = {October},
year = {2021},
pages = {3396-3405}
}
An overview of the current analysis framework will be available as a QMSKI abstract
@InProceedings{Sahu_2022_QMSKI,
author = {Sahu, Pranjal and Greer, Hastings and Xu, Zhenlin and Shen, Zhengyang and Bonaretti, Serena and McCormick, Matt and Niethammer, Marc},3
title = {Reproducible Workflow for Visualization and Analysis of OsteoArthritis Abnormality Progression},
booktitle = {Proceedings of the International Workshop on Quantitative Musculoskeletal Imaging (QMSKI)},
year = {2022}
}
Results obtained by the OAI Analysis pipeline can be found in this manuscript [paper]
@article{huang2022dadp,
title={DADP: Dynamic abnormality detection and progression for longitudinal knee magnetic resonance images from the Osteoarthritis Initiative},
author={Huang, Chao and Xu, Zhenlin and Shen, Zhengyang and Luo, Tianyou and Li, Tengfei and Nissman, Daniel and Nelson, Amanda and Golightly, Yvonne and Niethammer, Marc and Zhu, Hongtu},
journal={Medical Image Analysis},
pages={102343},
year={2022},
publisher={Elsevier}
}
This work was developed with support in part from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) under award numbers 1R44AR074375 and 1R01AR072013.