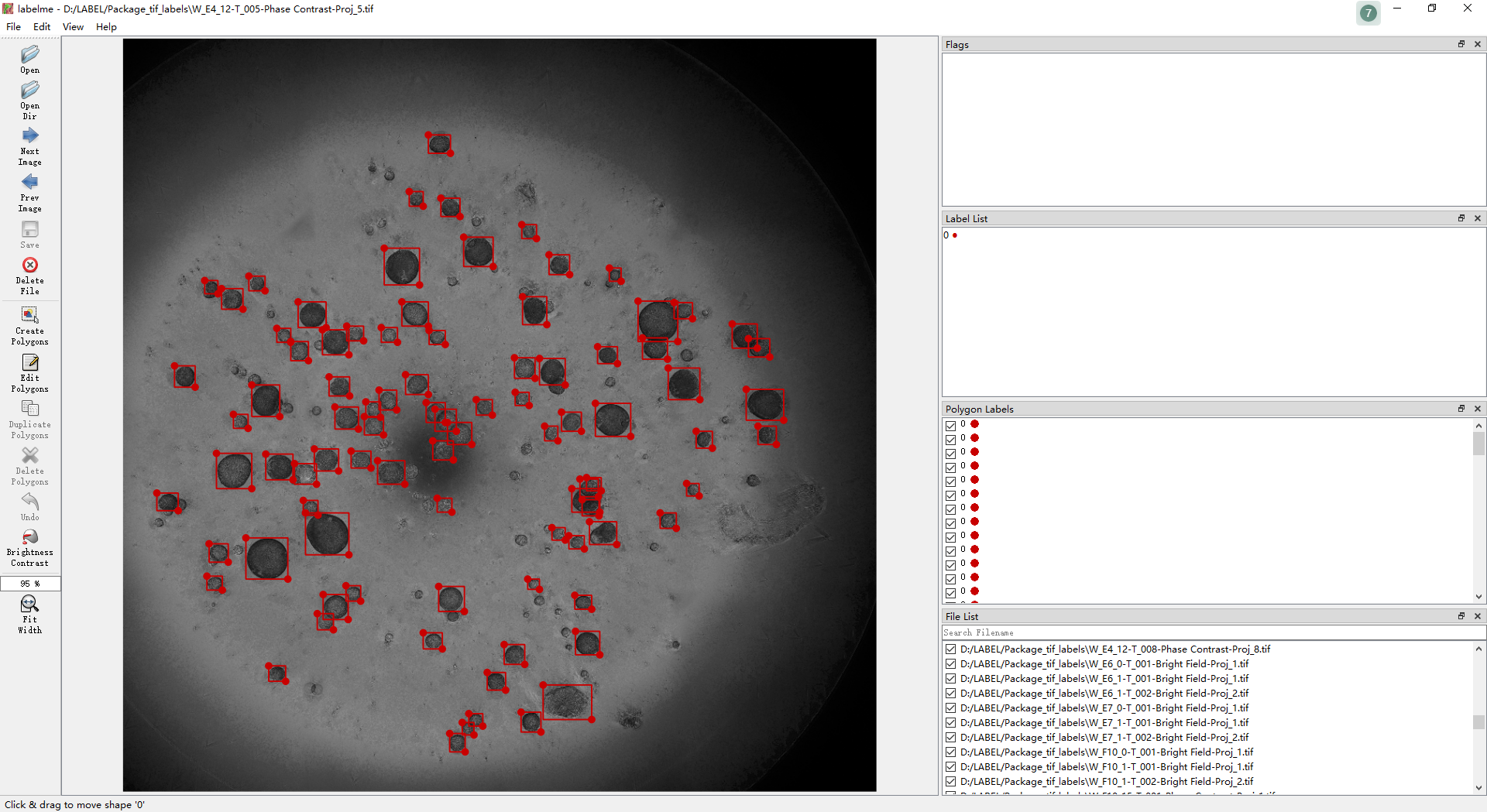
This is a open high-throughput organoid dataset for tracking. All organoids derive from mouse liver which is cultured in 96-well plate. We take one image by BioTek Cytation5 cell imaging multi-mode reader for each well with the interval of 24 hours.
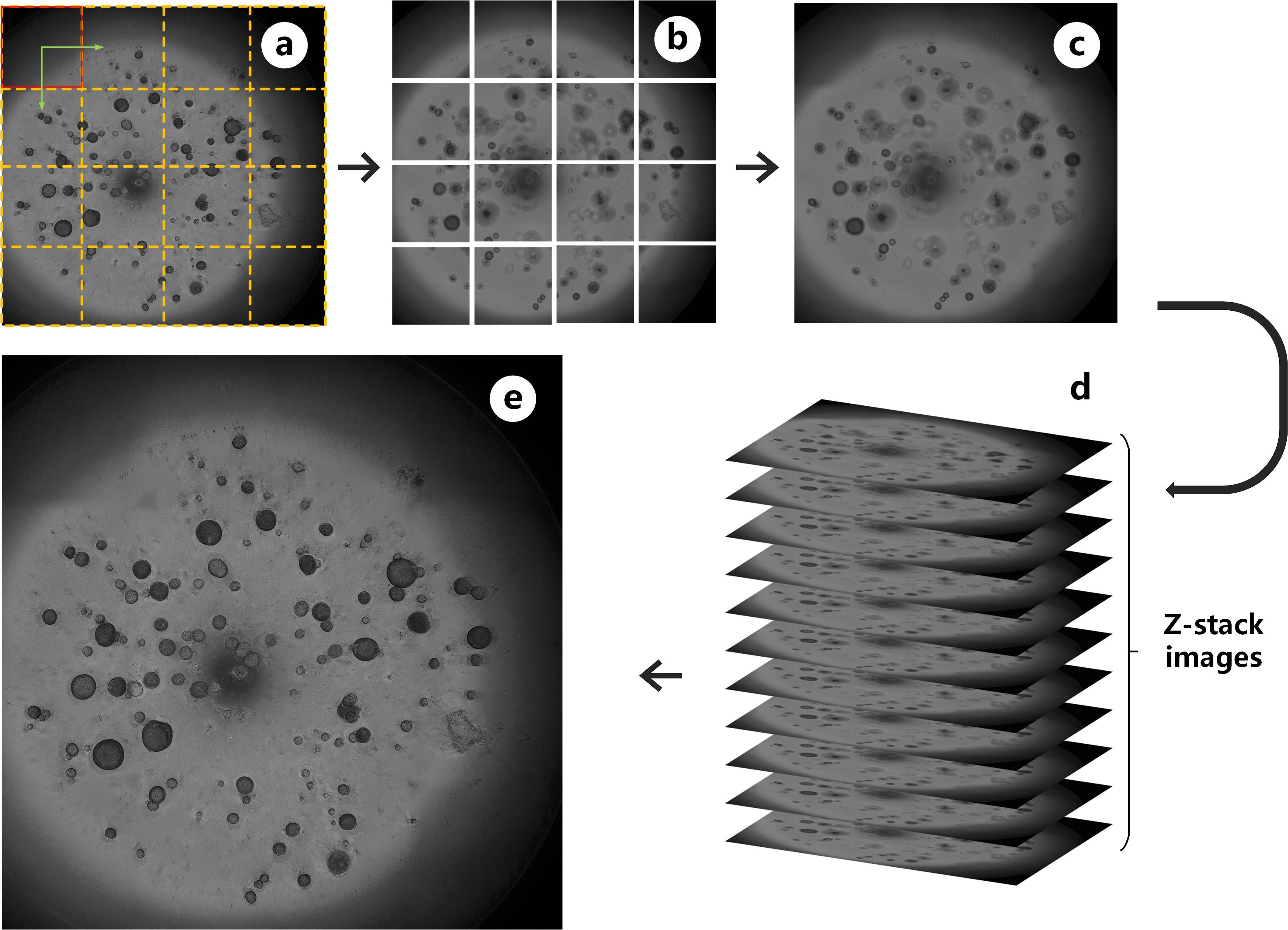 Pipeline for capturing high-throughout organoid images. The dash frame in (a) are FOVs for scanning. Image patches in (b) are montages captured from 16 FOVs. (c) shows the stitched images from specific focus plane. 32 z-stack images in (d) are projected to generate (e).
Pipeline for capturing high-throughout organoid images. The dash frame in (a) are FOVs for scanning. Image patches in (b) are montages captured from 16 FOVs. (c) shows the stitched images from specific focus plane. 32 z-stack images in (d) are projected to generate (e).
Organoid
├─Detection
│ ├─W_C10_0 # well name
│ │ Proj_1.tif # annotations
│ │ Proj_1.json # raw high-throughput organoid image
│ ├─W_C10_1
│ ├─W_C10_9
│ ├─......
│
└─Tracking
├─W_E4_12 # well name
│ Proj_1.json # annotations
│ Proj_1.tif # raw high-throughput organoid image
│ Proj_2.json
│ Proj_2.tif
│ Proj_3.json
│ Proj_3.tif
│ ......
│
├─W_F10_15
├─W_F10_3
├─ ......All annotation are created by labelme software (version 4.5.7). The json file for each image can be parsed by labelme directly and get the detailed information. In addition, we can also use the json package to read the annotation information.
The json file contains two kinds of key information: First,the position of each qualified organoid; Second, the identity of each qualified organoid.
{
"version": "4.5.6",
"flags": {},
"shapes": [
{
"label": "0",
"line_color": null,
"fill_color": null,
"points": [
[
746.0,
402.0
],
[
801.0,
455.0
]
],
"shape_type": "rectangle",
"flags": {}
}
// other shape
],
"imagePath": "XXXXXXXXXX.tif",
"imageData": "......",
"imageHeight": 9122,
"imageWidth": 9163
}We provide three ways to access this dataset.
mail to xsbian@stu.xmu.edu.cn
The data are captured by Accurate International Biotechnology (GZ) Co., Ltd, Nanfang Hospital and Southern Medical University. All annotation are created by team of Fujian Key Laboratory of Sensing and Computing for Smart City, Xiamen University. The data is available for free to researchers for non-commercial use.