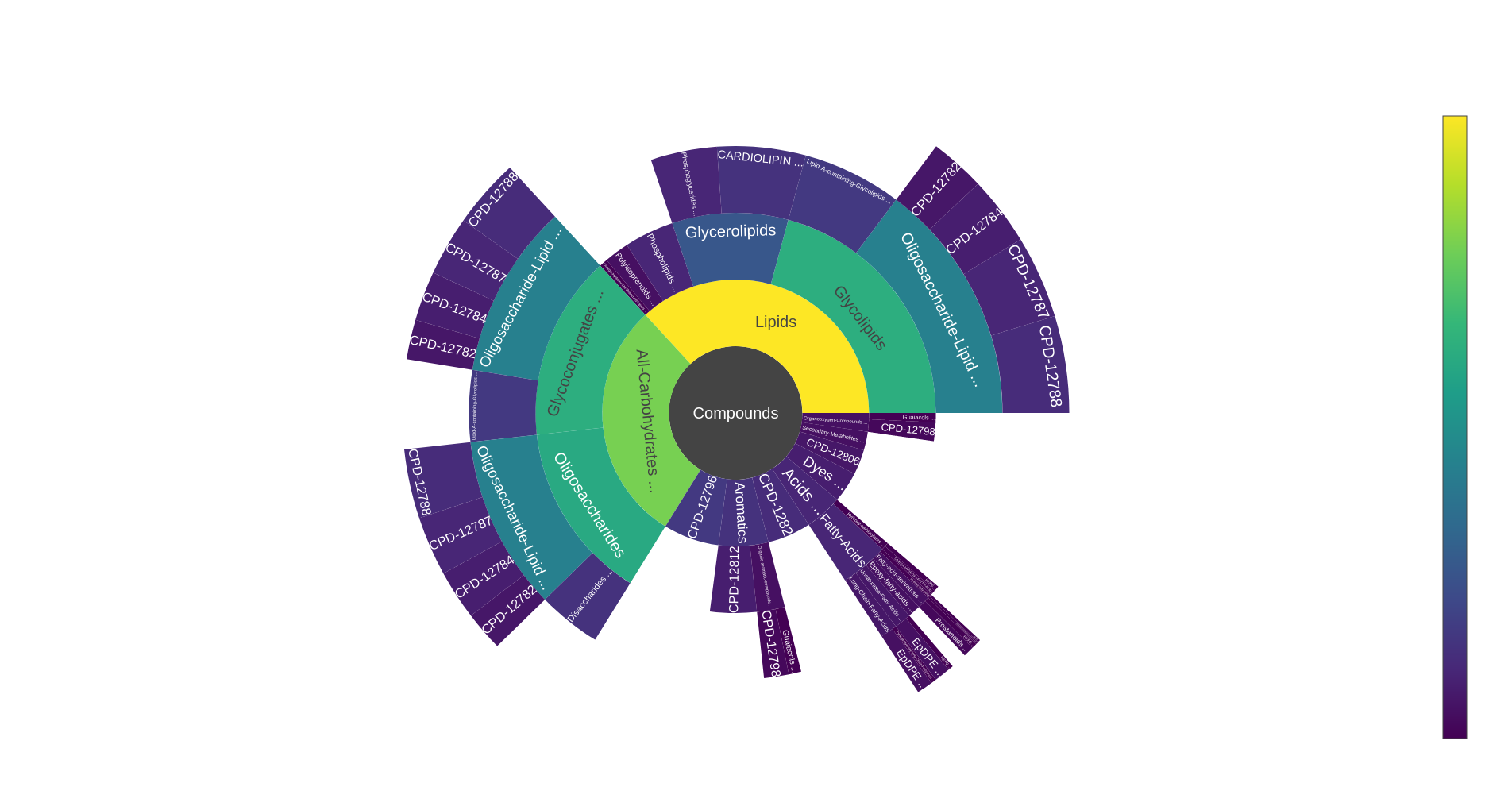
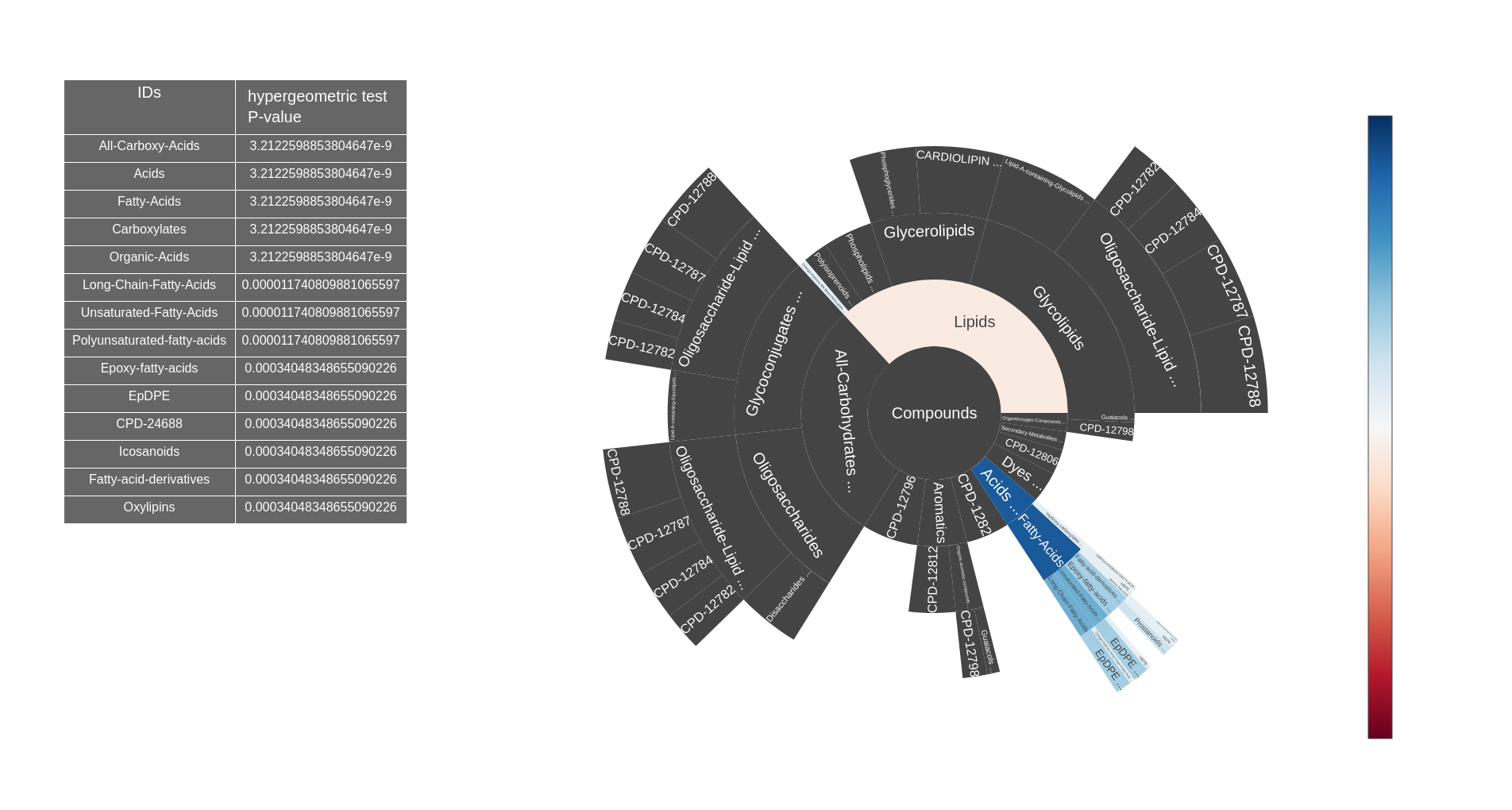
Sunburst visualisation of an ontology representing classes of sets of metabolic objects
Python 3.10 recommended
Requirements from requirements.txt
- numpy>=1.26.1
- plotly>=5.17.0
- scipy>=1.11.3
- SPARQLWrapper>=2.0.0
- pandas>=1.5.3
Need Apache Jena Fuseki SPARQL server for ChEBI and GO requests and their OWL files.
- Download Apache Jena Fuseki : https://jena.apache.org/download/index.cgi
- Download ChEBI ontology : https://ftp.ebi.ac.uk/pub/databases/chebi/ontology/ (chebi.owl or chebi_lite.owl)
- Download GO ontology : https://geneontology.org/docs/download-ontology/ (go.owl)
pip install ontosunburst
Inside the cloned repository :
pip install -r requirements.txt
pip install -e .
Execute followed bash script to launch server.
#!/bin/bash
FUSEKI_PATH=/path/to/apache-jena-fuseki-x.x.x
CHEBI_PATH=/path/to//chebi_lite.owl
${FUSEKI_PATH}/fuseki-server --file=${CHEBI_PATH} /chebi#!/bin/bash
FUSEKI_PATH=/path/to/apache-jena-fuseki-x.x.x
GO_PATH=/path/to/go.owl
${FUSEKI_PATH}/fuseki-server --file=${GO_PATH} /goWith local files :
- MetaCyc (compounds, reactions, pathways)
- EC (EC-numbers)
- KEGG Ontology (modules, pathways, ko, ko_transporter, metabolite, metabolite_lipid)
With SPARQL server :
- ChEBI (chebi roles)
- Gene Ontology (<1000 go terms recommended in the interest set)
Personal ontology possible :
- Define all the ontology classes relationship in
a dictionary
{class: [parent classes]} - Define the root : unique class with no parents
- Topology (1 set + 1 optional reference set) : displays proportion (number of occurrences) representation of all classes
- Enrichment (1 set + 1 reference set) : displays enrichment analysis significance of a set according to a reference set of metabolic objects
View full documentation here : https://github.com/AuReMe/Ontosunburst/wiki