Generalized Coarse Graining (GCG) enables the recursive coarse graining of discrete energy landscapes into gradient basins, funnels, super funnels, ..., using a gradient-neighbor-based aggregation directive.
The Perl5 script gcgBarriers.pl computes the gradient-based coarse graining of a
given energy landscape encoding as produced by the
barriers tool when enabling
full rate computation activated by its --rates argument.
Usage: perl gcgBarriers.pl <barriersOutput> <barriersRates>
The following call set exemplifies the usage of gcgBarriers.pl for the RNA secondary
structure energy landscape of riboswitch f07-a using the
Vienna RNA package.
Note, the calls are only viable for very short sequences since the whole
accessible structure space / energy landscape is considered.
# define RNA sequence of interest
RNA=GACCGGAAGGUCCGCCUUCC
# enumerate (and sort) all secondary structures (first by energy using structure string for tie breaking)
echo $RNA | RNAsubopt --deltaEnergy=99999 | sort -k 2,2n -k 1,1dr -S 2G | zip $RNA.RNAsubopt.zip -;
# run barriers to compute coarse graining level 1
unzip -p $RNA.RNAsubopt.zip | barriers --rates -G RNA -M noShift --bsize --max=999999 --minh=0 > $RNA.barriers.out;
# store rate matrix generated by barriers
mv rates.out $RNA.barriers.rates;
# cleanup obsolete barriers files
rm -f rates.bin tree.ps treeR.ps;
# compute generalized coarse grainings
perl gcgBarriers.pl $RNA.barriers.out $RNA.barriers.rates
The calls produce the output
level number
level states
0 2993
1 39
2 7
3 1
which states the number of macro-states on each coarse graining level.
Furthermore, the following files for each coarse graining level (LVL) > 1 are generated:
*.LVL.gradient: appends an additional column to thebarriersoutput file of the compressed level (LVL-1), which contains the gradient assignment for each local minimum*.LVL.barriers: a dummybarriersoutput file for the current level*.LVL.rates: the rates matrix for the current level
Given this data, you can plot the probability time series for the macro-states using
treekin.
# define variables for the following shell function
FILE="undef"; # base file name without extension (.barriers .rates)
OCID=0; # funnel of open chain state to be taken from according barriers output file
STEPS=1000000000; # maximal time (step) to compute
# shell function (tested in bash) to compute one plot via treekin
function runTreekin {
# ensure file naming for treekin call
ln -s $FILE.rates rates.out;
# output file prefix
OUTFILE=$FILE.treekin.m$OCID
# call treekin
treekin -m I --p0 $OCID=1 --t8=$STEPS < $FILE.barriers > $OUTFILE.out;
# cleanup temporary files
rm -f rates.out;
# generate output figure in pdf format using R
R --vanilla --silent -e "k <- read.table(\"$OUTFILE.out\", header=F, sep=\"\");pdf(\"$OUTFILE.pdf\");matplot(k[,1], k[,2:ncol(k)], main=\"$OUTFILE\", xlab=\"time (arbitrary units)\", ylab=\"state probability\", ylim=c(0,1), log=\"x\", type=\"l\");dev.off(); q();"
}
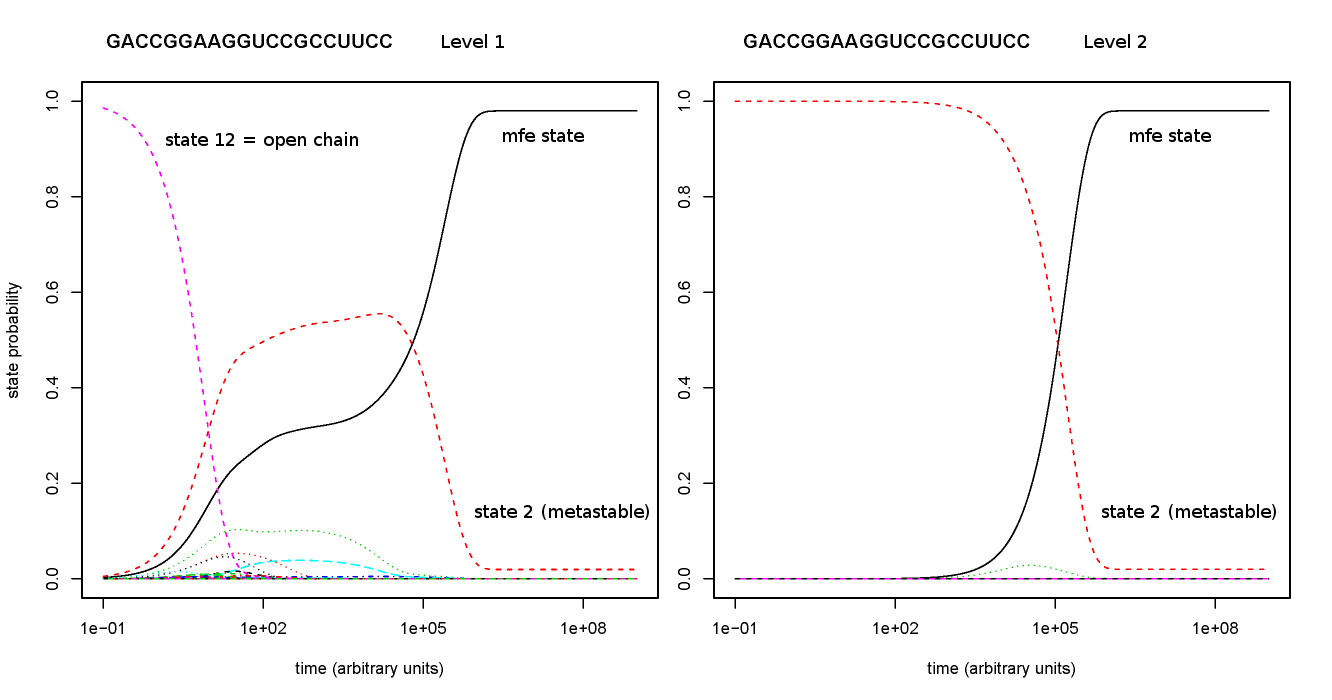
# define RNA sequence of interest
# e.g. riboswitch from https://doi.org/10.1002/bip.20761 Fig.7(left)
RNA=GACCGGAAGGUCCGCCUUCC
# level 1 plot
ln -s -f $RNA.barriers.out $RNA.barriers.out.1.barriers
ln -s -f $RNA.barriers.rates $RNA.barriers.out.1.rates
FILE=$RNA.barriers.out.1
OCID=12 # open chain state
runTreekin
# level 2 plot
FILE=$RNA.barriers.out.2
OCID=2 # gradient neighbor of open chain state from level 1
runTreekin
which will generate the following figures
Used program versions for the example output: ViennaRNA-2.4.4, barriers-1.6.0, treekin-0.3.1