Phonocardiogram Classification Using 1-Dimensional Inception Time Convolutional Neural Networks : George B. Moody PhysioNet Challenge 2022
This repository is associated with the paper: Phonocardiogram Classification Using 1-Dimensional Inception Time Convolutional Neural Networks
Full reference to the original paper :
B. -J. Singstad, A. M. Gitau, M. K. Johnsen, J. Ravn, L. A. Bongo and H. Schirmer, "Phonocardiogram Classification Using 1-Dimensional Inception Time Convolutional Neural Networks," 2022 Computing in Cardiology (CinC), Tampere, Finland, 2022, pp. 1-4, doi: 10.22489/CinC.2022.108.
- Markus Johnsen
- Johan Ravn
- Lars Ailo Bongo
- Henrik Schirmer
- Antony M. Gitau
- Bjørn-Jostein Singstad
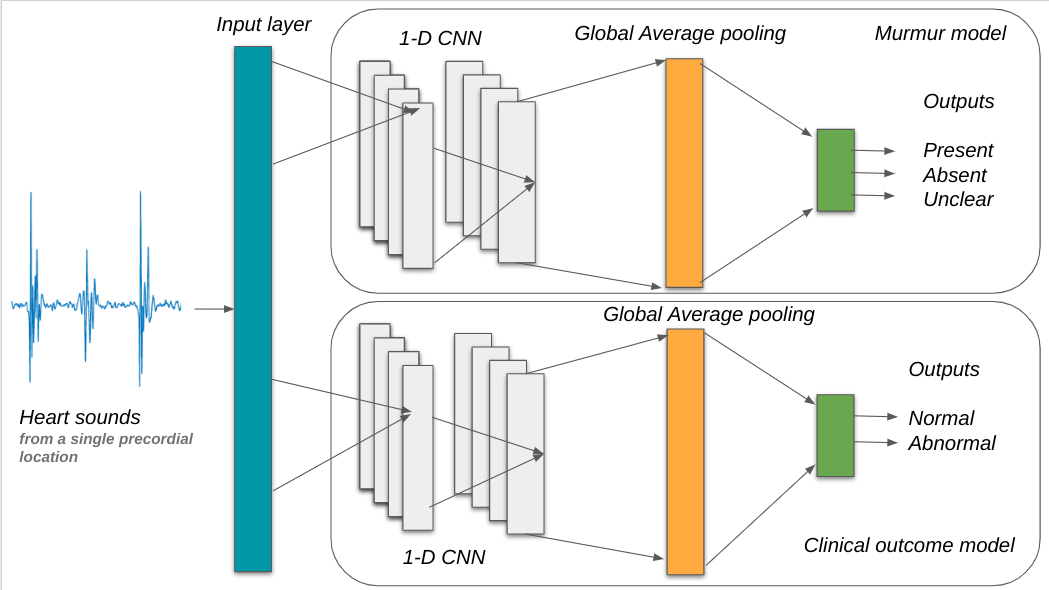
This repository contains the code for our contribution to the George B. Moody PhysioNet Challenge 2022. The objective of this challenge was to identify murmurs as present, absent or unclear and predict clinical outcome as normal or abnormal from phonocardiograms collected from multiple auscultation locations on the chest walls using digital stethoscope.
We trained and tested two 1-dimensional convolutional neural networks (CNN) on a PCG data set (5272 PCGs) from a pediatric population of 1568 individuals. One model predicted murmurs, while the other model predicted clinical outcomes. Both models were trained to give recording-wise predictions, while the final predictions were given for every patient (patient wise predictions).
Our team, Simulab, trained a clinical outcome classifier that achieved a challenge cost score of 8720 (ranked 1st out of 305 submissions) and the murmur classifier achieved a weighted
accuracy of 0.585 (ranked 182nd out of 305 submissions)
on the validation set.
This repository contains some scripts we can edit and some scripts provided by the organizers of George B. Moody PhysioNet Challenge 2022 which should NOT be edited
cross_validate.pyteam_code.py
evaluate_model.pyhelper_code.pyrun_model.pytrain_model.py
The repository also contains Jupyter Notebooks which makes easier to experiment with the data, models and other parameters. Some of the notebooks are designed to use in Google Colab in order to get access to GPUs and some notebooks are ok to run on your local computer.
5-fold cross-validaton.ipynb# contains code for cross-validating the model on the training setTrain and test model.ipynb# contains code training model a model on the whole training set, save the weights, then upload the wights and test the model on the training set and finaly evaluate the predictionspretrain model on 2015 dataset.ipynb# contains code for pretraing model on phonocardiogram data from PhysioNet Challenge 2016. returns a .h5 file
EDA-Phonocardiogram-dataset.ipynb# Exploratory data analysis of the dataset
This repository also contains the following files
.gitignoreLICENSEDockerfilerequirements.txt
The Dockerfile and the requirements.txt are very important as they are used to build the docker image when submitting our code to the challenge organizers.
The main data source is the CirCor DigiScope Dataset . When using the Jupyter Notebooks in Colab we need to download this dataset each time we start a new session.
To speed up the downloading we have created a Kaggle version of this dataset.
To download the dataset, you will have to sign up for a Kaggle account, get a kaggle.json file from you Kaggle profile (containing a API token) and upload it to the root folder in the temporary folder in Colab.
To pretrain the models we use the open dataset from PhysioNet Challenge 2016, also available on Kaggle.
numpy==1.21.6scipy==1.4.1scikit-learn==0.23.2joblib==0.17.0tensorflow == 2.8.2Cython==0.29.24pandas==1.3.2h5py==2.10.0tqdm==4.54.0
You can try it by running the following commands on the Challenge training sets. These commands should take a few minutes or less to run from start to finish on a recent personal computer.
For this example, we implemented a random forest classifier with several features. You can use a different classifier, features, and libraries for your entry. This simple example is designed not to perform well, so you should not use it as a baseline for your model's performance.
This code uses four main scripts, described below, to train and run a model for the 2022 Challenge.
You can install the dependencies for these scripts by creating a Docker image (see below) and running
pip install requirements.txt
You can train and run your model by running
python train_model.py training_data model
python run_model.py model test_data test_outputs
where training_data is a folder with the training data files, model is a folder for saving your model, test_data is a folder with the test data files (you can use the training data for debugging and cross-validation), and test_outputs is a folder for saving your model outputs. The 2022 Challenge website provides a training database with a description of the contents and structure of the data files.
You can evaluate your model by running
python evaluate_model.py labels outputs scores.csv class_scores.csv
where labels is a folder with labels for the data, such as the training database on the PhysioNet webpage; outputs is a folder containing files with your model's outputs for the data; scores.csv (optional) is a collection of scores for your model; and class_scores.csv (optional) is a collection of per-class scores for your model.
Docker and similar platforms allow you to containerize and package your code with specific dependencies so that you can run your code reliably in other computing environments and operating systems.
To guarantee that we can run your code, please install Docker, build a Docker image from your code, and run it on the training data. To quickly check your code for bugs, you may want to run it on a small subset of the training data.
If you have trouble running your code, then please try the follow steps to run the example code.
-
Create a folder
examplein your home directory with several subfolders.user@computer:~$ cd ~/ user@computer:~$ mkdir example user@computer:~$ cd example user@computer:~/example$ mkdir training_data test_data model test_outputs -
Download the training data from the Challenge website. Put some of the training data in
training_dataandtest_data. You can use some of the training data to check your code (and should perform cross-validation on the training data to evaluate your algorithm). -
Download or clone this repository in your terminal.
user@computer:~/example$ git clone https://github.com/physionetchallenges/python-classifier-2022.git -
Build a Docker image and run the example code in your terminal.
user@computer:~/example$ ls model python-classifier-2022 test_data test_outputs training_data user@computer:~/example$ cd python-classifier-2022/ user@computer:~/example/python-classifier-2022$ docker build -t image . Sending build context to Docker daemon [...]kB [...] Successfully tagged image:latest user@computer:~/example/python-classifier-2022$ docker run -it -v ~/example/model:/physionet/model -v ~/example/test_data:/physionet/test_data -v ~/example/test_outputs:/physionet/test_outputs -v ~/example/training_data:/physionet/training_data image bash root@[...]:/physionet# ls Dockerfile README.md test_outputs evaluate_model.py requirements.txt training_data helper_code.py team_code.py train_model.py LICENSE run_model.py root@[...]:/physionet# python train_model.py training_data model root@[...]:/physionet# python run_model.py model test_data test_outputs root@[...]:/physionet# python evaluate_model.py model test_data test_outputs [...] root@[...]:/physionet# exit Exit
| Model | Best parameters | Metrics | Training | Validation | Test |
|---|---|---|---|---|---|
| Murmur | Adam optimization | Weighted accuracy | 0.497 ± 0.083 | 0.585 | 0.593 |
| Weighted categorical cross entropy | Challenge metrics | 13158 ± 1283 | 8866 | 13134 | |
| 20 batch size | Accuracy | 0.446 ± 0.070 | 0.423 | 0.497 | |
| 30 epoch | F measure | 0.403 ± 0.055 | 0.384 | 0.398 | |
| Clinical | Adam optimization | Weighted accuracy | 0.713 ± 0.042 | 0.732 | 0.703 |
| 20 batch size | Challenge metrics | 12315 ± 903 | 8720 | 12419 | |
| 20 epoch | Accuracy | 0.51 ± 0.047 | 0.537 | 0.537 | |
| Weighted categorical cross entropy | F measure | 0.465 ± 0.061 | 0.530 | 0.503 |
@inproceedings{singstad_2022_phono,
title={Phonocardiogram classification using 1-dimensional inception time convolutional neural networks},
author={Singstad, Bj{\o}rn-Jostein and Gitau, Antony M and Johnsen, Markus Kreutzer and Ravn, Johan and Bongo, Lars Ailo and Schirmer, Henrik},
booktitle={2022 Computing in Cardiology (CinC)},
volume={498},
pages={1--4},
year={2022},
organization={IEEE}
}