Cluster Investigation & Virus Epidemiology Tool
- Install and update civet
- Input options
- Background data
- Usage
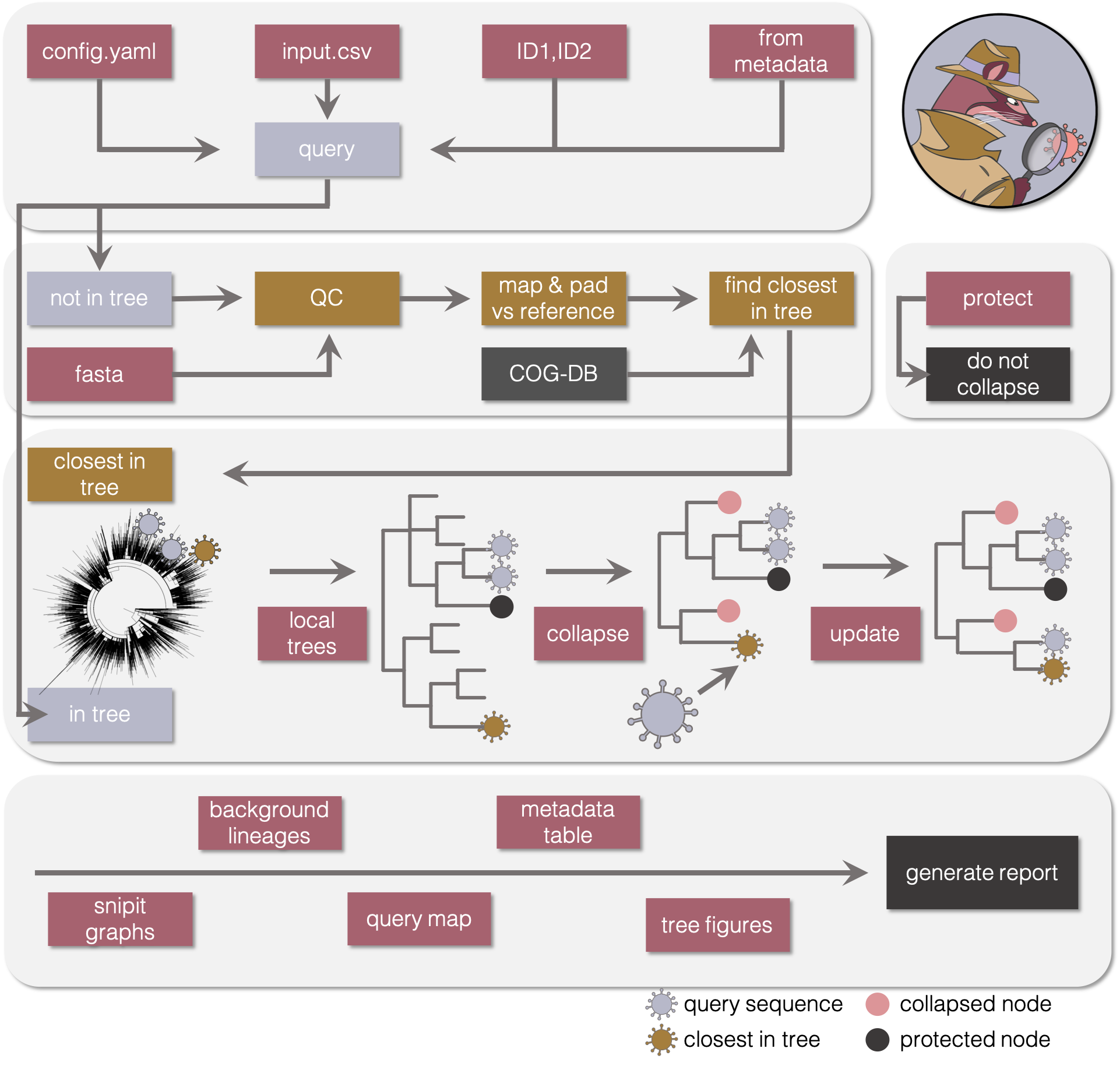
- Analysis pipeline
- Output
- Report options and descriptions
- Mapping options and usage instructions
- Adminstrative level 2 data
- Example report
- Contributors & acknowledgements
- Software versions
civet was created by Áine O'Toole & Verity Hill, Rambaut Group, Edinburgh University
Find out more information about civet at https://artic-network.github.io/civet/
civet is a tool developed with 'real-time' genomics in mind.
Using a background phylogeny, such as the large phylogeny available through the COG-UK infrastructure on CLIMB, civet will generate a report for a set of sequences of interest i.e. an outbreak investigation.
If the sequences are already on CLIMB and part of the large tree, civet will pull out the local context of those sequences, merging the smaller local trees as appropriate. If sequences haven't yet been incorporated into the large phylogeny, for instance if they have just been sequenced, civet will find the closest sequence in the large tree, pull the local tree of that sequence out and add your sequence in. The local trees then get collapsed to display in detail only the sequences of interest so as not to inform investigations beyond what was suggested by epidemiological data.
A fully customisable report is generated, summarising information about the sequences of interest. The tips of these trees can be coloured by any categorical trait present in the input csv, and additional fields added to the tip labels. Optional figures may be added to describe the local background of UK lineages and to map the query sequences using coordinates, again colourable by a custom trait.
__ __
____ |__|__ __ _____/ |_
/ ___\| \ \/ // __ \ __|
\ \___| |\ /\ ___/| |
\____/ __| \_/ \____/ __|
**** Cluster Investigation & Virus Epidemiology Tool ****
2.0
****************************************
Aine O'Toole & Verity Hill
Rambaut Group
Edinburgh University
Funding:
ARTIC Network
Wellcome Trust Collaborators Award
206298/Z/17/Z
COVID-19 Genomics UK Consortium
UK Department of Health and Social Care
UK Research and Innovation
ReservoirDOCs
European Research Council Consolidator Grant
ERC-2016-COG
Code contributors:
Ben Jackson gofasta
JT McCrone clusterfunk
Stefan Rooke local map
Andrew Rambaut jclusterfunk
Acknowledgements:
We thank the following for helpful suggestions,
comments, beta-testing, feature requests and
patience.
:nickloman: :mattloose:
:mattbashton: :tomconnor:
:rebeccadewar: :martinmchugh:
:richardmyers: :meerachand:
:samnicholls: :radpoplawski:
:davidaanensen: :benlindsey:
:jeffbarrett: :derekfairley:
:josephhughes: :davidrobertson:
:richardorton: :mattholden:
:ulfschaefer: :nataliegroves:
:nikosmanesis: :jaynaraghwani:
usage:
civet -i <config.yaml> [options]
civet -i input.csv [options]
civet -i ID1,IS2 [options]
civet -fm <column=match> [options]
input output options:
-i INPUT, --input INPUT
Input config file in yaml format, csv file (with
minimally an input_column header, Default=`name`) or
comma-separated id string with one or more query ids.
Example: `EDB3588,EDB3589`.
-fm [FROM_METADATA [FROM_METADATA ...]], --from-metadata [FROM_METADATA [FROM_METADATA ...]]
Generate a query from the metadata file supplied.
Define a search that will be used to pull out
sequences of interest from the large phylogeny. E.g.
-fm adm2=Edinburgh sample_date=2020-03-01:2020-04-01
-o OUTDIR, --outdir OUTDIR
Output directory. Default: current working directory
-f FASTA, --fasta FASTA
Optional fasta query.
--max-ambiguity MAX_AMBIGUITY
Maximum proportion of Ns allowed to attempt analysis.
Default: 0.5
--min-length MIN_LENGTH
Minimum query length allowed to attempt analysis.
Default: 10000
data source options:
-d DATADIR, --datadir DATADIR
Local directory that contains the data files. Default:
civet-cat
-m BACKGROUND_METADATA, --background-metadata BACKGROUND_METADATA
Custom metadata file that corresponds to the large
global tree/ alignment. Should have a column
`sequence_name`.
--background-tree BACKGROUND_TREE
Custom tree file in newick format, must match background metadata and background fasta file.
--background-sequences BACKGROUND_SEQUENCES
Custom background dataset in fasta format, must match background metadata and background tree file.
--CLIMB Indicates you're running CIVET from within CLIMB, uses
default paths in CLIMB to access data
-r, --remote Remotely access lineage trees from CLIMB
-uun UUN, --your-user-name UUN
Your CLIMB COG-UK username. Required if running with
--remote flag
--input-column INPUT_COLUMN
Column in input csv file to match with database.
Default: name
--data-column DATA_COLUMN
Option to search COG database for a different id type.
Default: COG-UK ID
report customisation:
-sc SEQUENCING_CENTRE, --sequencing-centre SEQUENCING_CENTRE
Customise report with logos from sequencing centre.
--display-name DISPLAY_NAME
Column in input csv file with display names for seqs.
Default: same as input column
--sample-date-column SAMPLE_DATE_COLUMN
Column in input csv with sampling date in it.
Default='sample_date'
--colour-by COLOUR_BY
Comma separated string of fields to display as
coloured dots rather than text in report trees.
Optionally add colour scheme eg adm1=viridis
--tree-fields TREE_FIELDS
Comma separated string of fields to display in the
trees in the report. Default: country
--label-fields LABEL_FIELDS
Comma separated string of fields to add to tree report
labels.
--date-fields DATE_FIELDS
Comma separated string of metadata headers containing
date information.
--node-summary NODE_SUMMARY
Column to summarise collapsed nodes by. Default =
Global lineage
--table-fields TABLE_FIELDS
Fields to include in the table produced in the report.
Query ID, name of sequence in tree and the local tree
it's found in will always be shown
--include-snp-table Include information about closest sequence in database
in table. Default is False
--no-snipit Don't run snipit graph
--include-bars Render barcharts in the output report
--omit-appendix Omit the appendix section. Default=False
--private remove adm2 references from background sequences.
Default=True
tree context options:
--distance DISTANCE Extraction from large tree radius. Default: 2
--up-distance UP_DISTANCE
Upstream distance to extract from large tree. Default:
2
--down-distance DOWN_DISTANCE
Downstream distance to extract from large tree.
Default: 2
--collapse-threshold COLLAPSE_THRESHOLD
Minimum number of nodes to collapse on. Default: 1
-p [PROTECT [PROTECT ...]], --protect [PROTECT [PROTECT ...]]
Protect nodes from collapse if they match the search
query in the metadata file supplied. E.g. -p
adm2=Edinburgh sample_date=2020-03-01:2020-04-01
map rendering options:
--local-lineages Contextualise the cluster lineages at local regional
scale. Requires at least one adm2 value in query csv.
--date-restriction Chose whether to date-restrict comparative sequences
at regional-scale.
--date-range-start DATE_RANGE_START
Define the start date from which sequences will COG
sequences will be used for local context. YYYY-MM-DD
format required.
--date-range-end DATE_RANGE_END
Define the end date from which sequences will COG
sequences will be used for local context. YYYY-MM-DD
format required.
--date-window DATE_WINDOW
Define the window +- either side of cluster sample
collection date-range. Default is 7 days.
--map-sequences Map the sequences themselves by adm2, coordinates or
otuer postcode.
--map-info MAP_INFO columns containing EITHER x and y coordinates as a
comma separated string OR outer postcodes for mapping
sequences OR Adm2
--input-crs INPUT_CRS
Coordinate reference system for sequence coordinates
--colour-map-by COLOUR_MAP_BY
Column to colour mapped sequences by
misc options:
-b, --launch-browser Optionally launch md viewer in the browser using grip
-c, --generate-config
Rather than running a civet report, just generate a
config file based on the command line arguments
provided
--tempdir TEMPDIR Specify where you want the temp stuff to go. Default:
$TMPDIR
--no-temp Output all intermediate files, for dev purposes.
--verbose Print lots of stuff to screen
-t THREADS, --threads THREADS
Number of threads
-v, --version show program's version number and exit
-h, --help