James S. Nagai1, Nils B. Leimkühler2, Michael T. Schaub 3, Rebekka K. Schneider4,5,6, Ivan G. Costa1*
1Institute for Computational Genomics, Faculty of Medicine, RWTH Aachen University, Aachen, 52074 Germany
2Department of Hematology and Stem Cell Transplantation, University Hospital Essen, Germany
3Department of Computer Science, RWTH Aachen University, Germany
4Department of Cell Biology, Institute for Biomedical Engineering, Faculty of Medicine,RWTH Aachen University, Pauwelsstrasse 30, 52074 Aachen, NRW, Germany
5Oncode Institute, Erasmus Medical Center, Rotterdam, 3015GD, the Netherlands
6Department of Hematology, Erasmus Medical Center, Rotterdam, 3015GD, the Netherlands
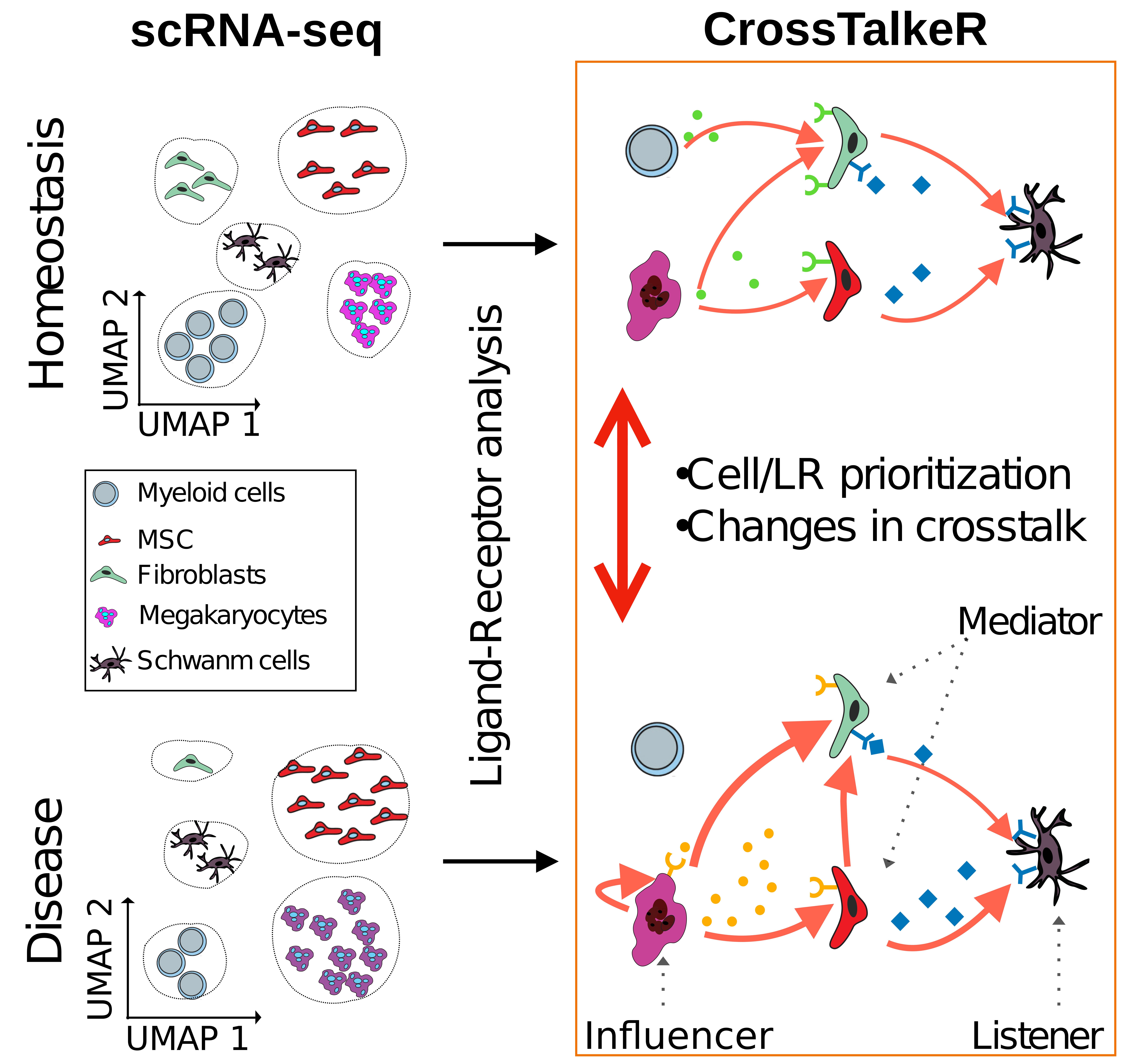
Motivation: Ligand-receptor (LR) analysis allows the characterization of cellular crosstalk from single cell RNA-seq data. However, current LR methods provide limited approaches for prioritization of cell types, ligands or receptors or characterizing changes in crosstalk between two biological conditions.
Results: pyCrossTalkeR is a framework for network analysis and visualisation of LR networks. pyCrossTalkeR identifies relevant ligands, receptors and cell types contributing to changes in cell communication when contrasting two biological states: disease vs. homeostasis. A case study on scRNA-seq of human myeloproliferative neoplasms reinforces the strengths of pyCrossTalkeR for characterisation of changes in cellular crosstalk in disease state.
You can install pyCrossTalkeR with the simple commands below:
pip install git+https://github.com/CostaLab/pyCrossTalkeR/
Note: Please avoid to use the following characters in celltype name: '$'
libudunits2-dev
libgdal-dev
gdal-bin
libproj-dev
proj-data
proj-bin
libgeos-dev
- Single and Comparative Reports
- Cell Cell Interaction visualization
- Sending and Receiving Cells Ranking
- CCI and GCI PCA ranking
- All measures and PC table
- PC1 and PC2 based barplot
- LR pair visualization plot can be done
[1] CrossTalkeR: Analysis and Visualisation of Ligand Receptor Networks link
[2] Heterogeneous bone-marrow stromal progenitors drive myelofibrosis via a druggable alarmin axis. link
[3] Comparison of Resources and Methods to infer Cell-Cell Communication from Single-cell RNA Data link