The HORTO-3DLM Dataset contains 3D LiDAR and GNSS/localization data for the purpose of 3D LiDAR-based place recognition, localization and mapping in horticultural environments.
- 25/02/2024 HORTO-3DLM v2.0 added GTJ34 and ON22 sequences
- 1/12/2024 HORTO-3DLM v1.0 Uploaded
Table Caption: Summary of all sequences
The Seq. column contains the sequence names. The M, Y, and C columns refer to the month, year, and country of recording, respectively. The total distance (Dist) of each sequence is measured in meters, while the scan size refers to the number of points in each scan.
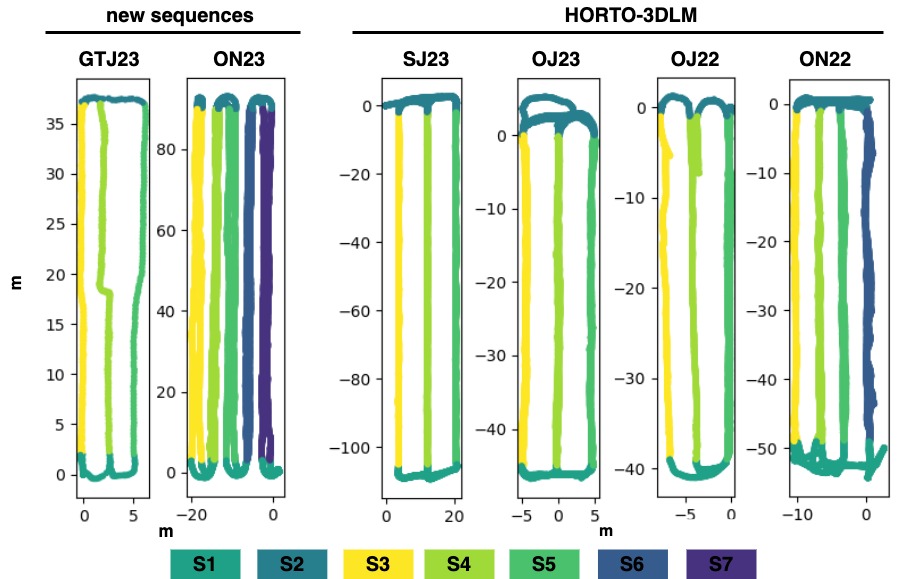
| Seq. | M | Y | C | Nº Scans | Nº Rows | Dist. [m] | Scan Size | Plantation Type |
|---|---|---|---|---|---|---|---|---|
| ON22 | Nov. | 2022 | UK | 7974 | 4 | 514 | 48k | Apple (open) |
| OJ22 | July | 2022 | UK | 4361 | 3 | 206 | 50k | Apple (open) |
| OJ23 | June | 2023 | UK | 7229 | 3 | 459 | 46k | Cherry (open) |
| SJ23 | June | 2023 | UK | 6389 | 3 | 742 | 48k | Strawberry (polytunnels) |
| ON23 | Nov. | 2023 | FR | 3086 | 5 | 966 | 105k | Apple (open) |
| GTJ23 | June | 2023 | PT | 661 | 3 | 202 | 60k | Tomato (greenhouse) |
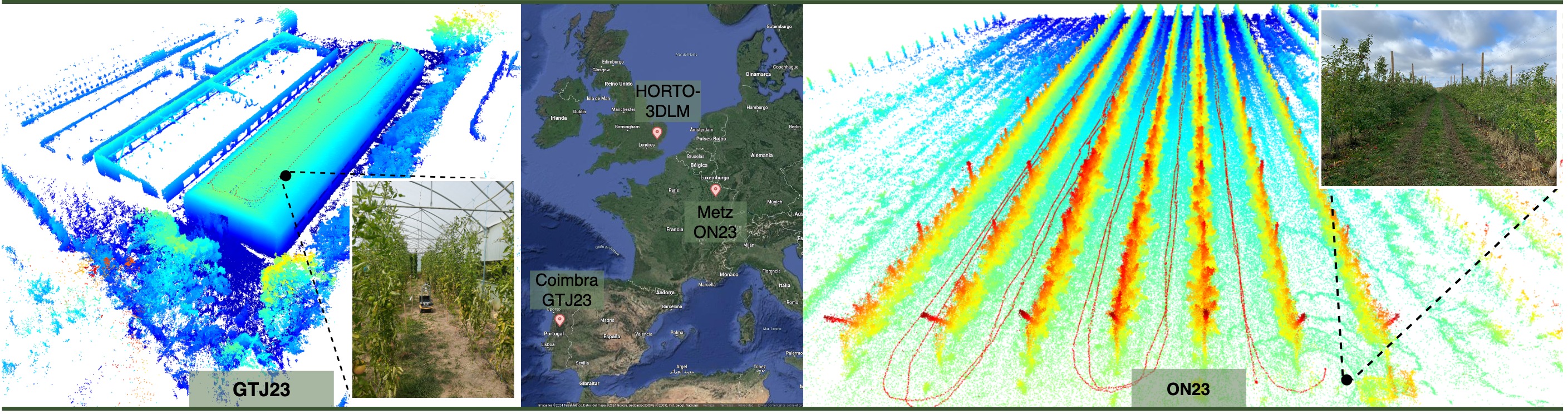
Sequence ON23 was recorded in November 2023, in an orchard in Metz, France, with an 16-beam Ouster 3D LiDAR and an SBG GNSS/INS system (without RTK) mounted on a Clearpath Husky mobile platform. To address the low LiDAR resolution, the original scans were merged to increase point density, resulting in sub-maps with approximately 100k points per sub-map. This operation reduced the original sequence from 25836 scans to 3086 sub-maps in total.
Sequence GTJ23 was recorded in June of 2023, in a tomato plantation within a greenhouse, in Coimbra, Portugal, with a 64-beam Ouster 3D LiDAR mounted on a Clearpath Jackal mobile platform. Due to signal interference caused by the greenhouse structure, the GNSS signal was unreliable. Therefore, the ground-truth positions were computed using a SLAM approach.
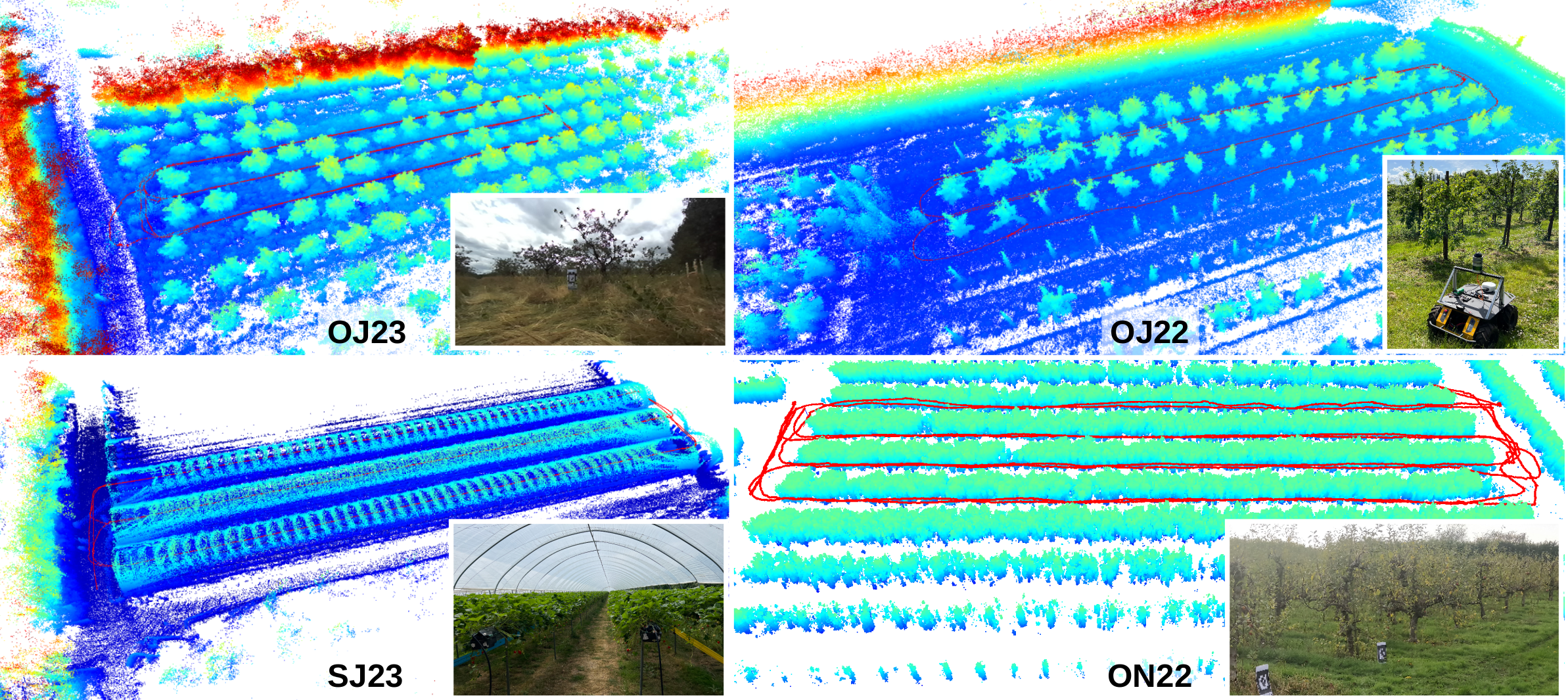
These sequences were recorded in England, UK, using a Clearpath Husky mobile robot equipped with a Velodyne VLP32 3D LiDAR (10Hz) and a ZED-F9P RTK-GPS (5Hz).
- OJ23: orchards,
- OJ22: orchards,
- SJ23: strawberries within polytunnels with a table-top growing system.
- ON22: orchards
HORTO-3DLM
├── GT23
├── OJ22
├── OJ23
├── ON22
├── ON23
└── SJ23
└── extracted
├── eval
| └──ground_truth_loop_range_10m.pkl
├── point_cloud
| ├── 0000000.bin
| ├── 0000001.bin
| ├── ...
| └── xxxxxxx.bin
├── triplet
| └── ground_truth_ar0.5m_nr10m_pr2m.pkl
├── extracted_info.txt
├── gps.txt
├── gps.kmz
├── gps_timestamp.txt
├── point_cloud_timestamp.txt
├── point_row_labels.pkl
├── positions.txt
├── positions_timestamp.txt
├── static_tf.txt
├── tf_poses.txt
└── tf_poses_timestamp.txt
- eval/ground_truth_loop_range_10m.pkl: pickle file with the ground truth data for evaluation(testing), containing anchors/positives indices for 10m range loops.
- point_cloud/: Folder with the point clouds.
- triplet/ground_truth_ar0.5m_nr10m_pr2m.pkl pickle file with the ground truth data for training, containing anchors, positives and negatives. The 0.5m between anchors; the positives were selected from within a 2m range; and negatives were selected from outside a 10m range.
- extracted_info.txt: some info from the extracted data;
- gps.txt: GNSS data in UTM reference system.
- gps.kmz: GNSS data for visualization on maps.
- gps_timestamp.txt: timestamps for GNSS data.
- point_cloud_timestamp.txt: timestamps for point cloud. row index 00 -> points cloud 00000.bin.
- point_row_labels.pkl: segment labels for each position data.
- positions.txt: position data with (x,y,z), used in place recognition for ground truth.
- positions_timestamp.txt: timestamps for position data
- static_tf.txt: static tf between base_link and LiDAR
- tf_poses.txt: pose data in the format of transformation matrix flattened to fit one file row.
- tf_poses_timestamp.txt: timestamp of the tf_pose.txt data.
@article{barros2023orchnet,
title={ORCHNet: A Robust Global Feature Aggregation approach for 3D LiDAR-based Place recognition in Orchards},
author={Barros, T and Garrote, L and Conde, P and Coombes, MJ and Liu, C and Premebida, C and Nunes, UJ},
journal={arXiv preprint arXiv:2303.00477},
year={2023}
}
- Add information
- Add Sensor TFs
- Add Dataset structure