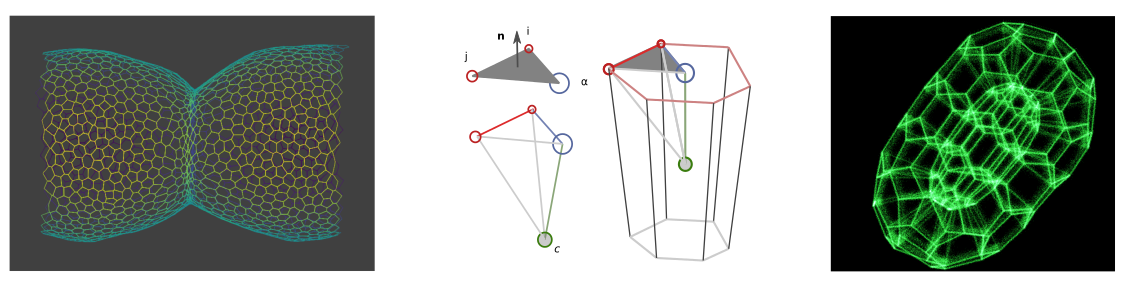
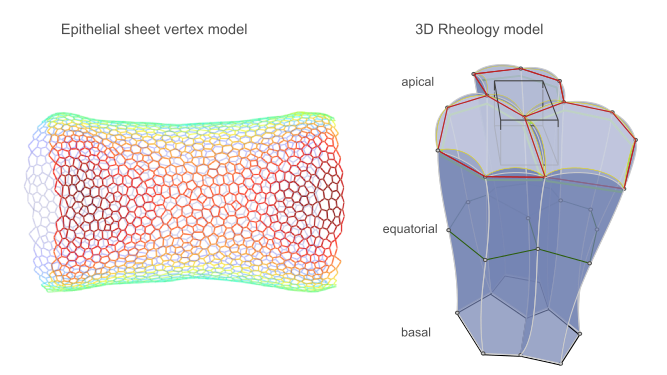
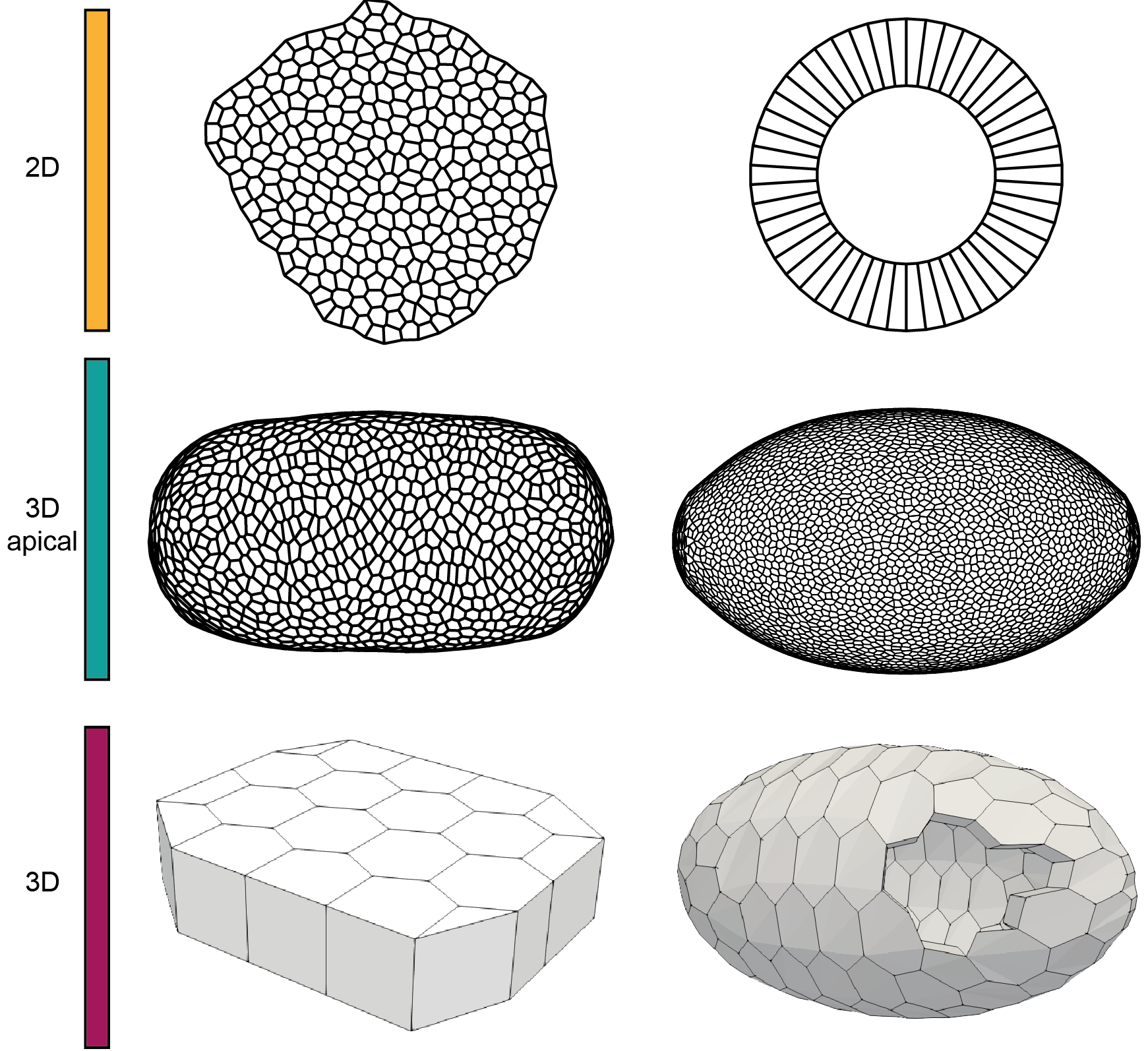
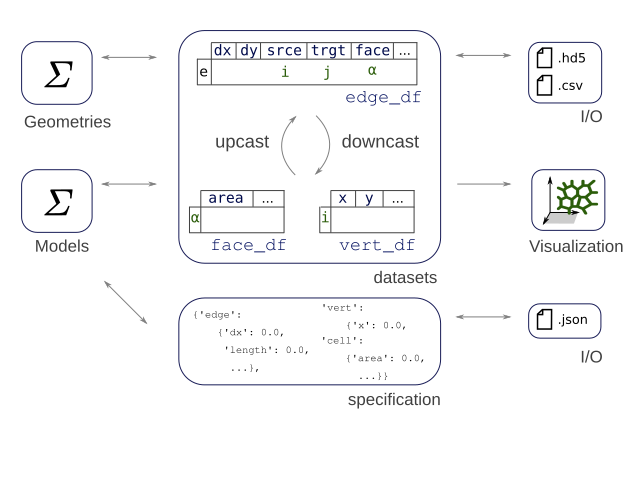
Dear tyssue user or prospective user, I come with good bearings.
This commit to the **new default branch main is the official start of the 1.0 release. It is long overdue and there is a lot of house keeping.
- fix CI w/ github actions
- Readthedocs with mkdoc
- Tests
- Notebook tests
- Merge PRs #263 #274
- Tests
- Check Polarization notebooks
| Name | Downloads | Version | Platforms |
|---|---|---|---|
 |
 |
 |
 |
| Coverage | Doc | CHAT |
|---|---|---|
| ZENODO DOI | JOSS DOI |
|---|---|
The tyssue library seeks to provide a unified interface to implement
biomechanical models of living tissues.
It's main focus is on vertex based epithelium models.
The first model implemented is the one described in Monier et al.1. It is an example of a vertex model, where the interactions are only evaluated on the apical surface sheet of the epithelium. The second class of models is still at a stage. They implement a description of the tissue's rheology, within a dissipation function formalism.
tyssue allows to model different geometries in 2D and in 3D. Presentation of the different geometries and how to create them are shown in this notebook.
Each biological question, be it in morphogenesis or cancer studies is unique, and requires tweeking of the models developed by the physicists. Most of the modelling software follow an architecture based on a core C++ engine with a combinaison of markup or scripting capacities to run specific simulation.
In tyssue, we rather try to expose an API that simplifies the
building of tissue models and running simulations, while keeping the
possibilities as open as possible.
Separate structure, geometry and models
We seek to have a design as modular as possible, to allow the same epithelium mesh to be fed to different physical models.
Accessible, easy to use data structures
The core of the tyssue library rests on two structures: a set of
pandas DataFrame holding the tissue geometry and associated data,
and nested dictionaries holding the model parameters, variables and
default values.
The API thus defines an Epithelium class. An instance of this class
is a container for the datasets and the specifications, and implements
methods to manipulate indexing of the dataframes to ease calculations.
The mesh structure is heavily inspired by CGAL Linear Cell Complexes, most importantly, in the case of a 2D vertex sheet for example, each junction edge between the cells is "splited" between two oriented half edges.
## Core object
from tyssue import Sheet
## Simple 2D geometry
from tyssue import PlanarGeometry
## Visualisation (matplotlib based)
from tyssue.draw import sheet_view
sheet = Sheet.planar_sheet_2d('basic2D', nx=6, ny=7,
distx=1, disty=1)
PlanarGeometry.update_all(sheet)
sheet.sanitize()
fig, ax = sheet_view(sheet)- The documentation is now browsable on tyssue.io
- The old documentation is still browsable online here
- Introduction notebooks are available here.
Thanks to @kephale, there is a napari plugin to visualise tyssue simulation output. You can find it here.
- No collision in 2D (use effector
Repulsion) - Add new geometry : 2D lateral geometry
- Add mean calculation in
Epithelium - Add
sheetconversion tomeshfromMeshio - Allow hole in 2D sheet
- Fix some visualisation
You are welcome to participate in the development of Tyssue.
What is planned for the future of Tyssue?
- Solve collision in 2.5D & 3D
- Use ZARR instead of HDF5 as base file format
- Upgrade geometry creation
- Compound geometry to form complex shape
- Use biological image as blueprint
- Add rheology model
- Switch to github-action for continuous integration
tyssue@framaliste.org - https://framalistes.org/sympa/info/tyssue
Subscribe ➙ https://framalistes.org/sympa/subscribe/tyssue Unsubscribe ➙ https://framalistes.org/sympa/sigrequest/tyssue
- Bertrand Caré - @bcare
- Cyprien Gay - @cypriengay
- Guillaume Gay (maintainer) - @glyg
- Hadrien Mary - @hadim
- François Molino
- Magali Suzanne
- Sophie Theis - @sophietheis
As all the dependencies are already completely supported in python 3.x, we won't be maintaining a python 2.x version, because it's time to move on...
Show dependencies
- CGAL > 4.7
- Python >= 3.6
- numpy
- scipy
- matplotlib
- pandas
- pytables
- jupyter
- notebook
- quantities
- ipywidgets
- pythreejs
- ipyvolume
- vispy
- pytest
- coverage
- pytest-cov
You can install the library with the conda package manager
conda install -c conda-forge tyssueYou can also install tyssue from PyPi, this is a CGAL-less version (pure python), lacking some features:
python -m pip install --user --upgrade tyssue
See INSTALL.md for a step by step install, including the necessary python environment.
If you find tyssue useful please cite this repository using its DOI as follows:
Theis, Suzanne, Gay, (2021). Tyssue: an epithelium simulation library. Journal of Open Source Software, 6(62), 2973 doi:https://doi.org/10.21105/joss.02973
Zenodo doi:10.5281/zenodo.4817609
If tyssue has made a substantial contribution to your work, please edit this file and open a PR.
Lou Y, Rupprecht JF, Theis S, Hiraiwa T, and Saunders TE. Curvature-induced cell rearrangements in biological tissues. Phys. Rev. Lett. 2023. doi: 10.1103/PhysRevLett.130.108401
Rahman T, Peters F and Wan LQ. Cell Jamming Regulates Epithelial Chiral Morphogenesis. J Biomech. 2023. doi: 10.1016/j.jbiomech.2023.111435
Fiorentino J and Scialdone A. The role of cell geometry and cell-cell communication in gradient sensing. PLoS Comput Biol. 2022 doi: 10.1371/journal.pcbi.1009552 Related repository: https://github.com/ScialdoneLab/2DLEGI
Courcoubetis G, Xu C, Nuzhdin SV, Haas S. Avalanches during epithelial tissue growth; Uniform Growth and a drosophila eye disc model, PLoS Comput Biol 2022 doi: 10.1371/journal.pcbi.1009952
Martin E, Theis S, Gay G, Monier B, Rouvière C, Suzanne M. Arp2/3-dependent mechanical control of morphogenetic robustness in an inherently challenging environment. Dev Cell. 2021 doi: 10.1016/j.devcel.2021.01.005 Related repository: https://github.com/suzannelab/polarity
Gracia M, Theis S, Proag A, Gay G, Benassayag C, Suzanne M. Mechanical impact of epithelial-mesenchymal transition on epithelial morphogenesis in Drosophila. Nat Commun. 2019 doi: 10.1038/s41467-019-10720-0 Related repository: https://github.com/suzannelab/invagination
Monier B, Gettings M, Gay G, Mangeat T, Schott S, Guarner A, Suzanne M. Apico-basal forces exerted by apoptotic cells drive epithelium folding. Nature. 2015 doi: 10.1038/nature14152 Related repository: https://github.com/glyg/leg-joint
Since version 0.3, this project is distributed under the terms of the General Public Licence.
Versions 2.4 and earlier were distributed under the Mozilla Public Licence.
If GPL licencing is too restrictive for your intended usage, please contact the maintainer.
Please note that this repository is participating in a study into sustainability of open source projects. Data will be gathered about this repository for approximately the next 12 months, starting from June 2021.
Data collected will include number of contributors, number of PRs, time taken to close/merge these PRs, and issues closed.
For more information, please visit our informational page or download our participant information sheet.
1 : Monier B, Gettings M, Gay G, Mangeat T, Schott S, Guarner A, Suzanne M. Apico-basal forces exerted by apoptotic cells drive epithelium folding. Nature. 2015 doi: 10.1038/nature14152 ↩