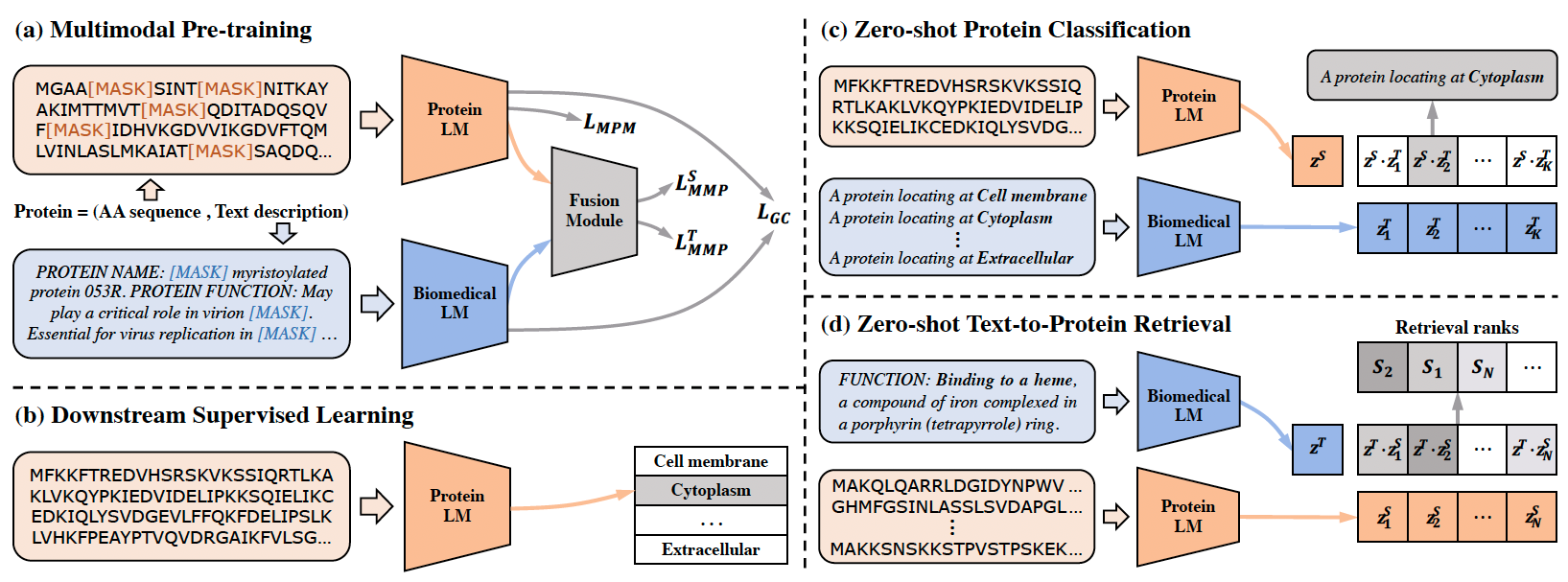
ProtST is an advanced pretraining framework for protein sequence understanding and prediction, as introduced in our ICML2023 oral paper. It is designed to enhance protein sequence pre-training and understanding by integrating protein functions and other important properties through biomedical texts.
The effectiveness and superiority of ProtST-induced PLMs over previous ones are demonstrated on diverse representation learning downstream tasks and zero-shot predictions. It also enables functional protein retrieval from large-scale databases even without any function annotation, as illustrated below.
You may install the dependencies of TorchProtein and ProtST as below. Generally, they work with Python 3.7/3.8 and PyTorch version >= 1.8.0.
conda create -n protein python=3.9
conda activate protein
conda install pytorch==2.0.0 pytorch-cuda=11.7 -c pytorch -c nvidia
conda install torchdrug pytorch-sparse pytorch-scatter pytorch-cluster -c pytorch -c pyg -c milagraph
conda install scikit-learn pandas decorator ipython networkx tqdm matplotlib -y
conda install fair-esm transformers easydict pyyaml lmdb -c conda-forge| Model | Config | Ckpt |
|---|---|---|
| ProtST-ESM-1b | config | ckpt |
| ProtST-ESM-2 | config | ckpt |
| ProtST-ProtBert | config | ckpt |
To reproduce all the experiments in ProtST, we provide all the necessary configuration files at config/.../*.yaml, which are categorized by the dataset, model architecture, and hyperparameters. When running experiments, we specify the configuration file with an argument --config and all the required arguments marked by {{ }} in that configuration file.
Note that all the datasets will be automatically downloaded in the code. But if you are using clusters without Internet connection, please run python ./script/prepare_all_datasets.py to cache datasets in advance.
By default, we pretrain 3 different PLM backbones (ESM-1b, ESM2 and ProtBert) using 4 V100 GPUs with the following command. Note that we have the choice of using two versions of text encoders: PebMedBert trained with only abstracts PebMedBert-abs and PebMedBert trained with full papers PebMedBert-full.
alias python4proc='python -m torch.distributed.launch --nproc_per_node=4'
# pretrain ESM-1b
python4proc script/run_pretrain.py --config ./config/pretrain/pretrain_esm.yaml --protein_model ESM-1b --text_model PubMedBERT-abs
# pretrain ESM-2
python4proc script/run_pretrain.py --config ./config/pretrain/pretrain_esm.yaml --protein_model ESM-2-650M --text_model PubMedBERT-abs
# pretrain ProtBert
python4proc script/run_pretrain.py --config ./config/pretrain/pretrain_protbert.yaml --text_model PubMedBERT-abs
For representation learning, we verify our pre-trained multimodal PLMs on 11 standard benchmarks for protein localization prediction, fitness landscape prediction and protein function annotation, under both fix-encoder learning and full-model tuning settings.
We label the pretrained checkpoints as PRETRAIN_CHECKPOINT. For different PLM backbone, the corresponding configuration files are in ./config/downstream_task/.../*.yaml. We give a demonstration for ProtST-enhanced ESM-1b.
For binary localization prediction, you can run as below to perform fix-encoder learning and full-model tuning, respectively:
# fix-encoder learning
python4proc ./script/run_downstream.py --config ./config/downstream_task/PretrainESM/localization_fix.yaml --checkpoint $PRETRAIN_CHECKPOINT --dataset BinaryLocalization --num_class 2
# full-model tuning
python4proc ./script/run_downstream.py --config ./config/downstream_task/PretrainESM/localization_tune.yaml --checkpoint $PRETRAIN_CHECKPOINT --dataset BinaryLocalization --num_class 2
Note that, subcellular localization can be performed in the similar way (please see ./config for details).
For Beta-Lactamase fitness prediction, you can run as below to perform fix-encoder learning and full-model tuning, respectively:
# fix-encoder learning
python4proc ./script/run_downstream.py --config ./config/downstream_task/PretrainESM/fitness_fix.yaml --checkpoint $PRETRAIN_CHECKPOINT --dataset BetaLactamase --batch_size 32
# full-model tuning
python4proc ./script/run_downstream.py --config ./config/downstream_task/PretrainESM/fitness_tune.yaml --checkpoint $PRETRAIN_CHECKPOINT --dataset BetaLactamase --batch_size 6
Note that, Fluorescence, Stability, AAV and Thermostability prediction can be performed in the similar way (please see ./config for details).
For Enzyme Commission (EC) number prediction, you can run as below to perform full-model tuning:
python4proc ./script/run_downstream.py --config ./config/downstream_task/PretrainESM/annotation_tune.yaml --checkpoint $PRETRAIN_CHECKPOINT --dataset td_datasets.EnzymeCommission --branch null
Note that, the Gene Ontology (GO) term prediction at Molecular Function (MF), Biological Process (BP) and Cellular Component (CC) branches can be performed in the similar way (please see ./config for details).
ProtST supports zero-shot protein classification, where it does not require any labeled protein. This is achieved by comparing representation similarities between a query protein and all labels, thanks to the aligned representation space of protein sequences and label descriptions in ProtST.
We demonstrate on zero-shot subcellular localization prediction and zero-shot reaction classification with ProtST-enhanced ESM-1b. We have also explored different prompt templates and description fields as listed in ./data/zero_shot_classification/.
# Subcellular Localization Prediction
python ./script/run_zero_shot.py --config ./config/zero_shot/PretrainESM/zero_shot.yaml --checkpoint $PRETRAIN_CHECKPOINT --prompt_label ./data/zero_shot_classification/subloc_template.tsv --dataset SubcellularLocalization --field "['name']"
# Reaction Classification
python ./script/run_zero_shot.py --config ./config/zero_shot/PretrainESM/zero_shot.yaml --checkpoint $PRETRAIN_CHECKPOINT --prompt_label ./data/zero_shot_classification/reaction_name.tsv --dataset Reaction --field "['name']"
ProtST-induced zero-shot classifiers have better data efficiency against various few-shot and non-parametric classifiers. You can run these baselines as below:
# few-shot classifiers
## Subcellular Localization Prediction
python ./script/run_few_shot.py --config ./config/few_shot/PretrainESM/few_shot.yaml --dataset SubcellularLocalization --num_class 10 --checkpoint $PRETRAIN_CHECKPOINT
## Reaction Classification
python ./script/run_few_shot.py --config ./config/few_shot/PretrainESM/few_shot.yaml --dataset Reaction --num_class 384 --checkpoint $PRETRAIN_CHECKPOINT
# non-parametric few-shot classifiers
## Subcellular Localization Prediction
python ./script/run_few_shot_nonparam.py --config ./config/few_shot/PretrainESM/few_shot.yaml --dataset SubcellularLocalization --num_class 10 --checkpoint $PRETRAIN_CHECKPOINT
## Reaction Classification
python ./script/run_few_shot_nonparam.py --config ./config/few_shot/PretrainESM/few_shot.yaml --dataset Reaction --num_class 384 --checkpoint $PRETRAIN_CHECKPOINT
We also show that ProtST-based zero-shot predictor can enhance the performance of supervised learning models via ensemble. We use the following scripts to do ensembles, where SUPERVISED_CHECKPOINT refers to the checkpoints obtained by supervised learning on downstream tasks.
## Subcellular Localization Prediction
python ./script/run_supervised_with_zero.py -sc ./config/downstream_task/PretrainESM/localization_fix.yaml -zc ./config/zero_shot/zero_shot.yaml --dataset SubcellularLocalization --num_class 10 --prompt_label ./data/zero_shot_classification/subloc_name.tsv --field "['name']" --checkpoint $PRETRAIN_CHECKPOINT --supervised_checkpoint $SUPERVISED_CHECKPOINT
## Reaction Classification
python ./script/run_supervised_with_zero.py -sc ./config/downstream_task/PretrainESM/reaction_tune.yaml -zc ./config/zero_shot/zero_shot.yaml --dataset Reaction --num_class 384 --prompt_label ./data/zero_shot_classification/reaction_name.tsv --field "['name']" --checkpoint $PRETRAIN_CHECKPOINT --supervised_checkpoint $SUPERVISED_CHECKPOINT
We illustrate the capability of ProtST-ESM-1b on retrieving functional proteins as below, where no function annotation is required:
python ./script/run_t2p_retrieval.py --config ./config/t2p_retrieval/go_mf.yaml --checkpoint $PRETRAIN_CHECKPOINT
This codebase is released under the Apache License 2.0 as in the LICENSE file.
If you find this project helpful, please cite our paper:
@article{xu2023protst,
title={ProtST: Multi-Modality Learning of Protein Sequences and Biomedical Texts},
author={Xu, Minghao and Yuan, Xinyu and Miret, Santiago and Tang, Jian},
journal={arXiv preprint arXiv:2301.12040},
year={2023}
}
For any questions or issues, open an issue or contact Minghao Xu (minghao.xu@mila.quebec) and Xinyu Yuan (xinyu.yuan@mila.quebec).