A tensorflow(keras) implementation of multitask learning package for semi-supervised learning on biological sequences
pip install seqlearnerThis repo is divided into 3 directories.
- The
seqlearnerdirectory contains all codes and jupyter notebooks. - The
seqlearner/data/directory is place where data is in. - The
seqlearner/results/directory contains all results plots, Logs and etc.
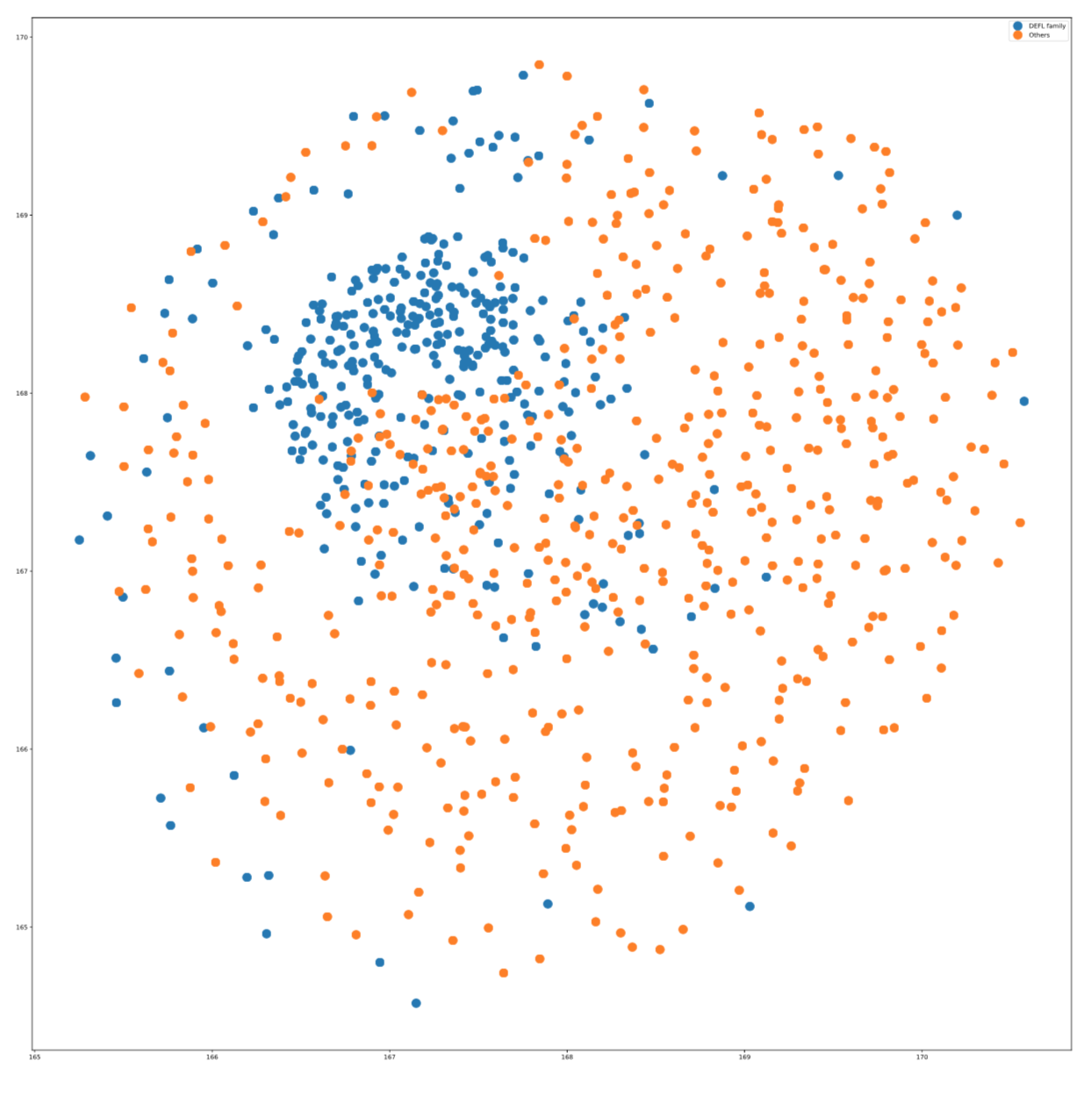
After Embedding the protein sequences with embedding methods, we provide some visualization for it. TSNE and UMAP have been used for visualizing embedding of 2 protein families to gather some evaluation about the embedding. With this evaluation we want to give some intuition about how well protein families are seperated via this embedding and the corresponding function.
Here is a simple example for calculating the embedding using Freq2Vec and visualize it via TSNE method.
First, you have to calculate and save the embedding via learner method.
freq2vec_embedding = mtl.embed(word_length=3, embedding="freq2vec", func="sum", emb_dim=25, gamma=0.1, epochs=100)after calculating the freq2vec embedding with 25 dimensions, we would like to visualize it via TSNE method.
visualize(method="TSNE", proportion=2.0)This will save a plot for you in seqlearner/results/ folder which the points are samples from 2 protein families which has the most samples in the dataset. Here is a sample plot.