Documents: https://bioinf.shenwei.me/unikmer/
unikmer is a toolkit for nucleic acid k-mer analysis,
providing functions
including set operation k-mers (sketch) optional with
TaxIds but without count information.
K-mers are either encoded (k<=32) or hashed (k<=64, using ntHash v1) into uint64,
and serialized in binary file with extension .unik.
TaxIds can be assigned when counting k-mers from genome sequences, and LCA (Lowest Common Ancestor) is computed during set opertions including computing union, intersecton, set difference, unique and repeated k-mers.
Related projects:
- kmers provides bit-packed k-mers methods for this tool.
- unik provides k-mer serialization methods for this tool.
- sketches provides generators/iterators for k-mer sketches (Minimizer, Scaled MinHash, Closed Syncmers).
- taxdump provides querying manipulations from NCBI Taxonomy taxdump files.
- Finding conserved regions in all genomes of a species.
- Finding species/strain-specific sequences for designing probes/primers.
-
Downloading executable binary files.
-
conda install -c bioconda unikmer
-
Counting
count Generate k-mers (sketch) from FASTA/Q sequences -
Information
info Information of binary files num Quickly inspect the number of k-mers in binary files -
Format conversion
view Read and output binary format to plain text dump Convert plain k-mer text to binary format encode Encode plain k-mer texts to integers decode Decode encoded integers to k-mer texts -
Set operations
concat Concatenate multiple binary files without removing duplicates inter Intersection of k-mers in multiple binary files common Find k-mers shared by most of the binary files union Union of k-mers in multiple binary files diff Set difference of k-mers in multiple binary files -
Split and merge
sort Sort k-mers to reduce the file size and accelerate downstream analysis split Split k-mers into sorted chunk files tsplit Split k-mers according to TaxId merge Merge k-mers from sorted chunk files -
Subset
head Extract the first N k-mers sample Sample k-mers from binary files grep Search k-mers from binary files filter Filter out low-complexity k-mers rfilter Filter k-mers by taxonomic rank -
Searching on genomes
locate Locate k-mers in genome map Mapping k-mers back to the genome and extract successive regions/subsequences -
Misc
autocompletion Generate shell autocompletion script version Print version information and check for update
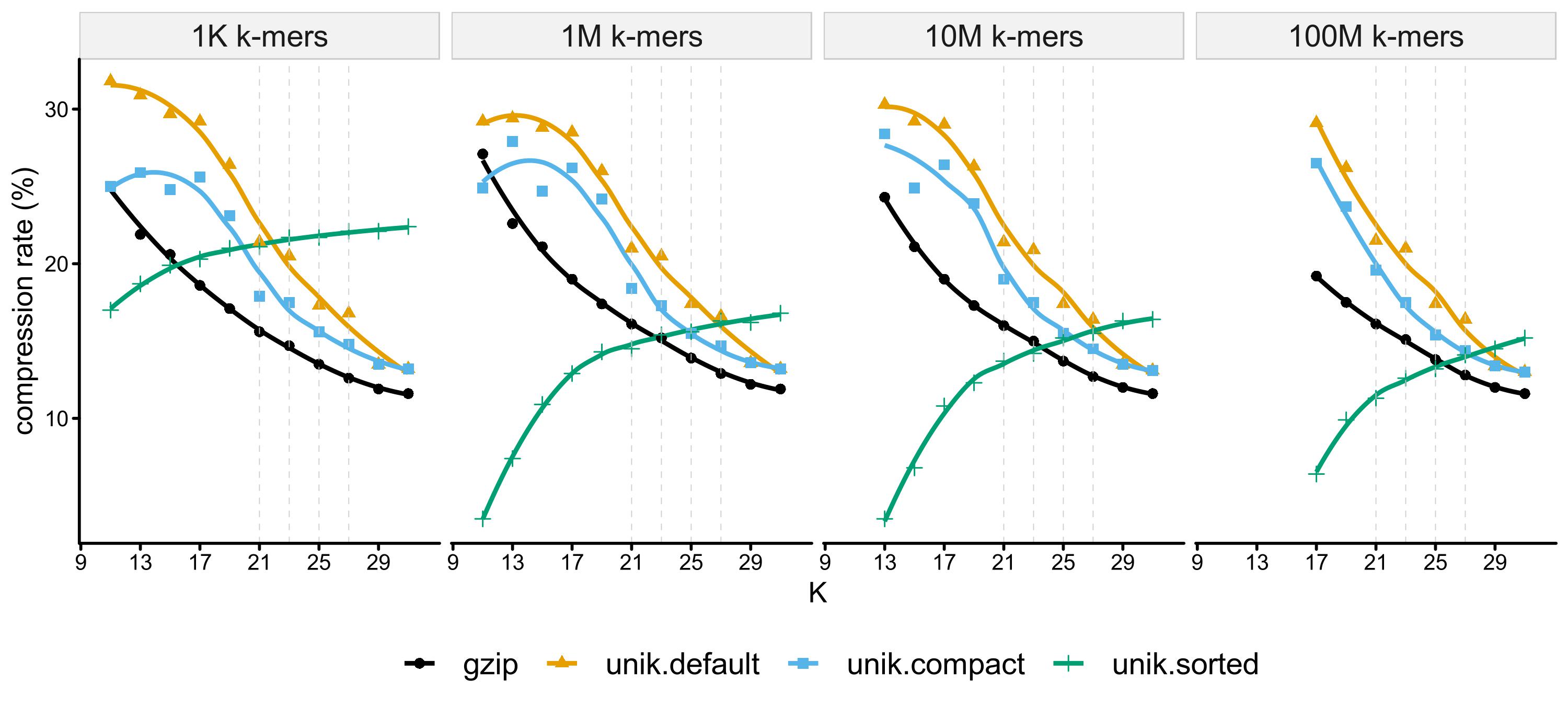
K-mers (represented in uint64 in RAM ) are serialized in 8-Byte
(or less Bytes for shorter k-mers in compact format,
or much less Bytes for sorted k-mers) arrays and
optionally compressed in gzip format with extension of .unik.
TaxIds are optionally stored next to k-mers with 4 or less bytes.
No TaxIds stored in this test.
| label | encoded-kmera | gzip-compressedb | compact-formatc | sortedd | comment |
|---|---|---|---|---|---|
plain |
plain text | ||||
gzip |
✔ | gzipped plain text | |||
unik.default |
✔ | ✔ | gzipped encoded k-mers in fixed-length byte array | ||
unik.compat |
✔ | ✔ | ✔ | gzipped encoded k-mers in shorter fixed-length byte array | |
unik.sorted |
✔ | ✔ | ✔ | gzipped sorted encoded k-mers |
- a One k-mer is encoded as
uint64and serialized in 8 Bytes. - b K-mers file is compressed in gzip format by default,
users can switch on global option
-C/--no-compressto output non-compressed file. - c One k-mer is encoded as
uint64and serialized in 8 Bytes by default. However few Bytes are needed for short k-mers, e.g., 4 Bytes are enough for 15-mers (30 bits). This makes the file more compact with smaller file size, controled by global option-c/--compact. - d One k-mer is encoded as
uint64, all k-mers are sorted and compressed using varint-GB algorithm. - In all test, flag
--canonicalis ON when runningunikmer count.
# memusg is for compute time and RAM usage: https://github.com/shenwei356/memusg
# counting (only keep the canonical k-mers and compact output)
# memusg -t unikmer count -k 23 Ecoli-IAI39.fasta.gz -o Ecoli-IAI39.fasta.gz.k23 --canonical --compact
$ memusg -t unikmer count -k 23 Ecoli-MG1655.fasta.gz -o Ecoli-MG1655.fasta.gz.k23 --canonical --compact
elapsed time: 0.897s
peak rss: 192.41 MB
# counting (only keep the canonical k-mers and sort k-mers)
# memusg -t unikmer count -k 23 Ecoli-IAI39.fasta.gz -o Ecoli-IAI39.fasta.gz.k23.sorted --canonical --sort
$ memusg -t unikmer count -k 23 Ecoli-MG1655.fasta.gz -o Ecoli-MG1655.fasta.gz.k23.sorted --canonical --sort
elapsed time: 1.136s
peak rss: 227.28 MB
# counting and assigning global TaxIds
$ unikmer count -k 23 -K -s Ecoli-IAI39.fasta.gz -o Ecoli-IAI39.fasta.gz.k23.sorted -t 585057
$ unikmer count -k 23 -K -s Ecoli-MG1655.fasta.gz -o Ecoli-MG1655.fasta.gz.k23.sorted -t 511145
$ unikmer count -k 23 -K -s A.muciniphila-ATCC_BAA-835.fasta.gz -o A.muciniphila-ATCC_BAA-835.fasta.gz.sorted -t 349741
# counting minimizer and ouputting in linear order
$ unikmer count -k 23 -W 5 -H -K -l A.muciniphila-ATCC_BAA-835.fasta.gz -o A.muciniphila-ATCC_BAA-835.fasta.gz.m
# view
$ unikmer view Ecoli-MG1655.fasta.gz.k23.sorted.unik --show-taxid | head -n 3
AAAAAAAAACCATCCAAATCTGG 511145
AAAAAAAAACCGCTAGTATATTC 511145
AAAAAAAAACCTGAAAAAAACGG 511145
# view (hashed k-mers needs original FASTA/Q file)
$ unikmer view --show-code --genome A.muciniphila-ATCC_BAA-835.fasta.gz A.muciniphila-ATCC_BAA-835.fasta.gz.m.unik | head -n 3
CATCCGCCATCTTTGGGGTGTCG 1210726578792
AGCGCAAAATCCCCAAACATGTA 2286899379883
AACTGATTTTTGATGATGACTCC 3542156397282
# find the positions of k-mers
$ unikmer locate -g A.muciniphila-ATCC_BAA-835.fasta.gz A.muciniphila-ATCC_BAA-835.fasta.gz.m.unik | head -n 5
NC_010655.1 2 25 ATCTTATAAAATAACCACATAAC 0 .
NC_010655.1 5 28 TTATAAAATAACCACATAACTTA 0 .
NC_010655.1 6 29 TATAAAATAACCACATAACTTAA 0 .
NC_010655.1 9 32 AAAATAACCACATAACTTAAAAA 0 .
NC_010655.1 13 36 TAACCACATAACTTAAAAAGAAT 0 .
# info
$ unikmer info *.unik -a -j 10
file k canonical hashed scaled include-taxid global-taxid sorted compact gzipped version number description
A.muciniphila-ATCC_BAA-835.fasta.gz.m.unik 23 ✓ ✓ ✕ ✕ ✕ ✕ ✓ v5.0 860,900
A.muciniphila-ATCC_BAA-835.fasta.gz.sorted.unik 23 ✓ ✕ ✕ ✕ 349741 ✓ ✕ ✓ v5.0 2,630,905
Ecoli-IAI39.fasta.gz.k23.sorted.unik 23 ✓ ✕ ✕ ✕ 585057 ✓ ✕ ✓ v5.0 4,902,266
Ecoli-IAI39.fasta.gz.k23.unik 23 ✓ ✕ ✕ ✕ ✕ ✓ ✓ v5.0 4,902,266
Ecoli-MG1655.fasta.gz.k23.sorted.unik 23 ✓ ✕ ✕ ✕ 511145 ✓ ✕ ✓ v5.0 4,546,632
Ecoli-MG1655.fasta.gz.k23.unik 23 ✓ ✕ ✕ ✕ ✕ ✓ ✓ v5.0 4,546,632
# concat
$ memusg -t unikmer concat *.k23.sorted.unik -o concat.k23 -c
elapsed time: 1.020s
peak rss: 25.86 MB
# union
$ memusg -t unikmer union *.k23.sorted.unik -o union.k23 -s
elapsed time: 3.991s
peak rss: 590.92 MB
# or sorting with limited memory.
# note that taxonomy database need some memory.
$ memusg -t unikmer sort *.k23.sorted.unik -o union2.k23 -u -m 1M
elapsed time: 3.538s
peak rss: 324.2 MB
$ unikmer view -t union.k23.unik | md5sum
4c038832209278840d4d75944b29219c -
$ unikmer view -t union2.k23.unik | md5sum
4c038832209278840d4d75944b29219c -
# duplicate k-mers
# memusg -t unikmer sort *.k23.sorted.unik -o dup.k23 -d -m 1M # limit memory usage
$ memusg -t unikmer sort *.k23.sorted.unik -o dup.k23 -d
elapsed time: 1.143s
peak rss: 240.18 MB
# intersection
$ memusg -t unikmer inter *.k23.sorted.unik -o inter.k23
elapsed time: 1.481s
peak rss: 399.94 MB
# difference
$ memusg -t unikmer diff -j 10 *.k23.sorted.unik -o diff.k23 -s
elapsed time: 0.793s
peak rss: 338.06 MB
$ ls -lh *.unik
-rw-r--r-- 1 shenwei shenwei 6.6M Sep 9 17:24 A.muciniphila-ATCC_BAA-835.fasta.gz.m.unik
-rw-r--r-- 1 shenwei shenwei 9.5M Sep 9 17:24 A.muciniphila-ATCC_BAA-835.fasta.gz.sorted.unik
-rw-r--r-- 1 shenwei shenwei 46M Sep 9 17:25 concat.k23.unik
-rw-r--r-- 1 shenwei shenwei 9.2M Sep 9 17:27 diff.k23.unik
-rw-r--r-- 1 shenwei shenwei 11M Sep 9 17:26 dup.k23.unik
-rw-r--r-- 1 shenwei shenwei 18M Sep 9 17:23 Ecoli-IAI39.fasta.gz.k23.sorted.unik
-rw-r--r-- 1 shenwei shenwei 29M Sep 9 17:24 Ecoli-IAI39.fasta.gz.k23.unik
-rw-r--r-- 1 shenwei shenwei 17M Sep 9 17:23 Ecoli-MG1655.fasta.gz.k23.sorted.unik
-rw-r--r-- 1 shenwei shenwei 27M Sep 9 17:25 Ecoli-MG1655.fasta.gz.k23.unik
-rw-r--r-- 1 shenwei shenwei 11M Sep 9 17:27 inter.k23.unik
-rw-r--r-- 1 shenwei shenwei 26M Sep 9 17:26 union2.k23.unik
-rw-r--r-- 1 shenwei shenwei 26M Sep 9 17:25 union.k23.unik
$ unikmer stats *.unik -a -j 10
file k canonical hashed scaled include-taxid global-taxid sorted compact gzipped version number description
A.muciniphila-ATCC_BAA-835.fasta.gz.m.unik 23 ✓ ✓ ✕ ✕ ✕ ✕ ✓ v5.0 860,900
A.muciniphila-ATCC_BAA-835.fasta.gz.sorted.unik 23 ✓ ✕ ✕ ✕ 349741 ✓ ✕ ✓ v5.0 2,630,905
concat.k23.unik 23 ✓ ✕ ✕ ✓ ✕ ✓ ✓ v5.0 -1
diff.k23.unik 23 ✓ ✕ ✕ ✓ ✓ ✕ ✓ v5.0 2,326,096
dup.k23.unik 23 ✓ ✕ ✕ ✓ ✓ ✕ ✓ v5.0 2,576,170
Ecoli-IAI39.fasta.gz.k23.sorted.unik 23 ✓ ✕ ✕ ✕ 585057 ✓ ✕ ✓ v5.0 4,902,266
Ecoli-IAI39.fasta.gz.k23.unik 23 ✓ ✕ ✕ ✕ ✕ ✓ ✓ v5.0 4,902,266
Ecoli-MG1655.fasta.gz.k23.sorted.unik 23 ✓ ✕ ✕ ✕ 511145 ✓ ✕ ✓ v5.0 4,546,632
Ecoli-MG1655.fasta.gz.k23.unik 23 ✓ ✕ ✕ ✕ ✕ ✓ ✓ v5.0 4,546,632
inter.k23.unik 23 ✓ ✕ ✕ ✓ ✓ ✕ ✓ v5.0 2,576,170
union2.k23.unik 23 ✓ ✕ ✕ ✓ ✓ ✕ ✓ v5.0 6,872,728
union.k23.unik 23 ✓ ✕ ✕ ✓ ✓ ✕ ✓ v5.0 6,872,728
# -----------------------------------------------------------------------------------------
# mapping k-mers to genome
seqkit seq Ecoli-IAI39.fasta.gz -o Ecoli-IAI39.fasta
g=Ecoli-IAI39.fasta
f=inter.k23.unik
# mapping k-mers back to the genome and extract successive regions/subsequences
unikmer map -g $g $f -a | more
# using bwa
# to fasta
unikmer view $f -a -o $f.fa.gz
# make index
bwa index $g; samtools faidx $g
ncpu=12
ls $f.fa.gz \
| rush -j 1 -v ref=$g -v j=$ncpu \
'bwa aln -o 0 -l 17 -k 0 -t {j} {ref} {} \
| bwa samse {ref} - {} \
| samtools view -bS > {}.bam; \
samtools sort -T {}.tmp -@ {j} {}.bam -o {}.sorted.bam; \
samtools index {}.sorted.bam; \
samtools flagstat {}.sorted.bam > {}.sorted.bam.flagstat; \
/bin/rm {}.bam '
Please open an issue to report bugs, propose new functions or ask for help.