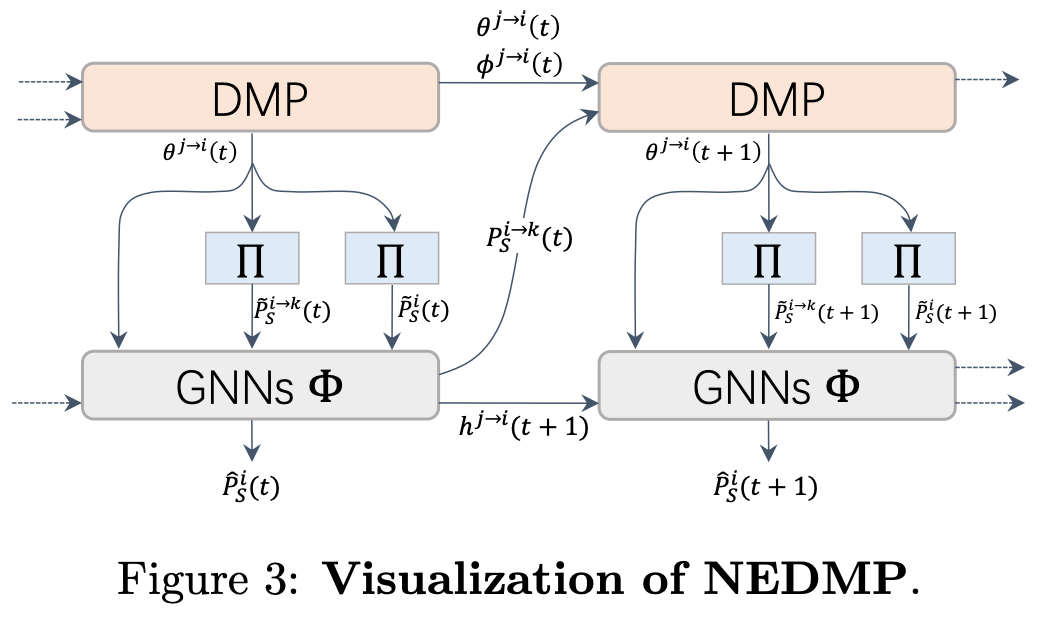
This is a PyTorch implementation of paper Neural Enhanced Dynamic Message Passing in AISTATS 2022.
Predicting stochastic spreading processes on complex networks is critical in epidemic control, opinion propagation, and viral marketing. We focus on the problem of inferring the time-dependent marginal probabilities of states for each node which collectively quantifies the spreading results. Dynamic Message Passing (DMP) has been developed as an efficient inference algorithm for several spreading models, and it is asymptotically exact on locally tree-like networks. However, DMP can struggle in diffusion networks with lots of local loops. We address this limitation by using Graph Neural Networks (GNN) to learn the dependency amongst messages implicitly. Specifically, we propose a hybrid model in which the GNN module runs jointly with DMP equations. The GNN module refines the aggregated messages in DMP iterations by learning from simulation data. We demonstrate numerically that after training, our model's inference accuracy substantially outperforms DMP in conditions of various network structure and dynamics parameters. Moreover, compared to pure data-driven models, the proposed hybrid model has a better generalization ability for out-of-training cases, profiting from the explicitly utilized dynamics priors in the hybrid model.
OS:
- Ubuntu
Install dependencies following:
conda install numpy -y
conda install networkx -y
pip install scipy==1.8.0
conda install pytorch==1.10.1 cudatoolkit=11.3 -c pytorch -c conda-forge
TORCH=1.10.1
CUDA=cu113
pip install torch-scatter -f https://pytorch-geometric.com/whl/torch-${TORCH}+${CUDA}.html
pip install torch-sparse -f https://pytorch-geometric.com/whl/torch-${TORCH}+${CUDA}.html
pip install torch-geometric
We provide bash scripts (at ./bashs) for generating all datasets used in our paper and running all experiments (as well as hyper parameters) conducted in our paper.
We implementate DMP using PyTorch at ./src/DMP/SIR.py, and the code for NEDMP and baseline GNN at src/model.
Notices: we use a binary file ./src/utils/simulation which is wrote in C and complied on Ubuntu to performe the simulations. Therefore, it may raise Error when you run ours codes on other operation systems.
The main script to generate simulation data is data_process/generate_train_data.py:
usage: generate_train_data.py [-h] [-data DATA_PATH] [-df DIFFUSION] [-ns NUM_SAMPLES] [-np NODE_PROB [NODE_PROB ...]] [-ep EDGE_PROB [EDGE_PROB ...]] [-ss SEED_SIZE]
[-cc CPU_CORE]
optional arguments:
-h, --help show this help message and exit
-data DATA_PATH, --data_path DATA_PATH
path to graphs
-df DIFFUSION, --diffusion DIFFUSION
The diffusion model
-ns NUM_SAMPLES, --num_samples NUM_SAMPLES
number of samples to be generated
-np NODE_PROB [NODE_PROB ...], --node_prob NODE_PROB [NODE_PROB ...]
the parameters range for node in diffusion model
-ep EDGE_PROB [EDGE_PROB ...], --edge_prob EDGE_PROB [EDGE_PROB ...]
the parameters range for edges in diffusion model
-ss SEED_SIZE, --seed_size SEED_SIZE
The maximum percent of nodes been set as nodes
-cc CPU_CORE, --cpu_core CPU_CORE
For example, by running:
python data_process/generate_train_data.py -df SIR -ns 200 -np 0.4 0.6 -ep 0.2 0.5 -ss 1 -data ./data/synthetic/tree
you can generate ns=200 independent SIR simulations in a date=tree graph each with ss=1 randomly selected seed node, and the propagation probibilities for each edge is randomly sampled from [0.2, 0.5], and the recover rate for each node is sample from [0.4, 0.6]. The resulting data will be saved to ./data/synthetic/tree/train_data/SIR_200.pkl
you can change the args -data to generate simulations on different graph strucutre.
Or you can simply run bash script ./bashs/generate_realnets_data.sh and ./bashs/generate_syn_data.sh to generate all datasets used for the fitting experiments.
Cause the space limitation, we only uploaded two simulation datasets: synthetic tree graph and real-world dolphins graph. You can train our model as well as the baseline model GNN on those toy datasets:
# dolphins
python train.py --model gnn --cuda_id 1 --diff SIR --data_path data/realnets/dolphins/train_data/SIR_150.pkl
python train.py --model nedmp --cuda_id 1 --diff SIR --data_path data/realnets/dolphins/train_data/SIR_150.pkl
# tree
python train.py --diff SIR --model nedmp --cuda_id -1 --data_path data/synthetic/tree/train_data/SIR_200.pkl
python train.py --diff SIR --model gnn --cuda_id -1 --data_path data/synthetic/tree/train_data/SIR_200.pkl
And by running ./bashs/train_syn_nedmp.sh, ./bashs/train_real_nedmp.sh,./bashs/train_syn_gnn.sh and ./bashs/train_real_gnn.sh, you can get all the results in the fitting experiments.
@InProceedings{pmlr-v151-gao22b,
title = { Neural Enhanced Dynamic Message Passing },
author = {Gao, Fei and Zhang, Jiang and Zhang, Yan},
booktitle = {Proceedings of The 25th International Conference on Artificial Intelligence and Statistics},
pages = {10471--10482},
year = {2022},
editor = {Camps-Valls, Gustau and Ruiz, Francisco J. R. and Valera, Isabel},
volume = {151},
series = {Proceedings of Machine Learning Research},
month = {28--30 Mar},
publisher = {PMLR},
pdf = {https://proceedings.mlr.press/v151/gao22b/gao22b.pdf},
url = {https://proceedings.mlr.press/v151/gao22b.html},
}