| output |
|---|
github_document |
JBrowseR is an R package that provides a simple and clean interface to JBrowse 2 for R users. Using JBrowseR, you can:
- Embed the JBrowse 2 genome browser in R Markdown documents and Shiny applications
- Deploy a genome browser directly from the R console to view your data
- Customize your genome browser to display your own data
You can install the released version of JBrowseR from CRAN with:
install.packages("JBrowseR")And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("gmod/JBrowseR")This line of code can be used to launch a genome browser from your R console:
library(JBrowseR)
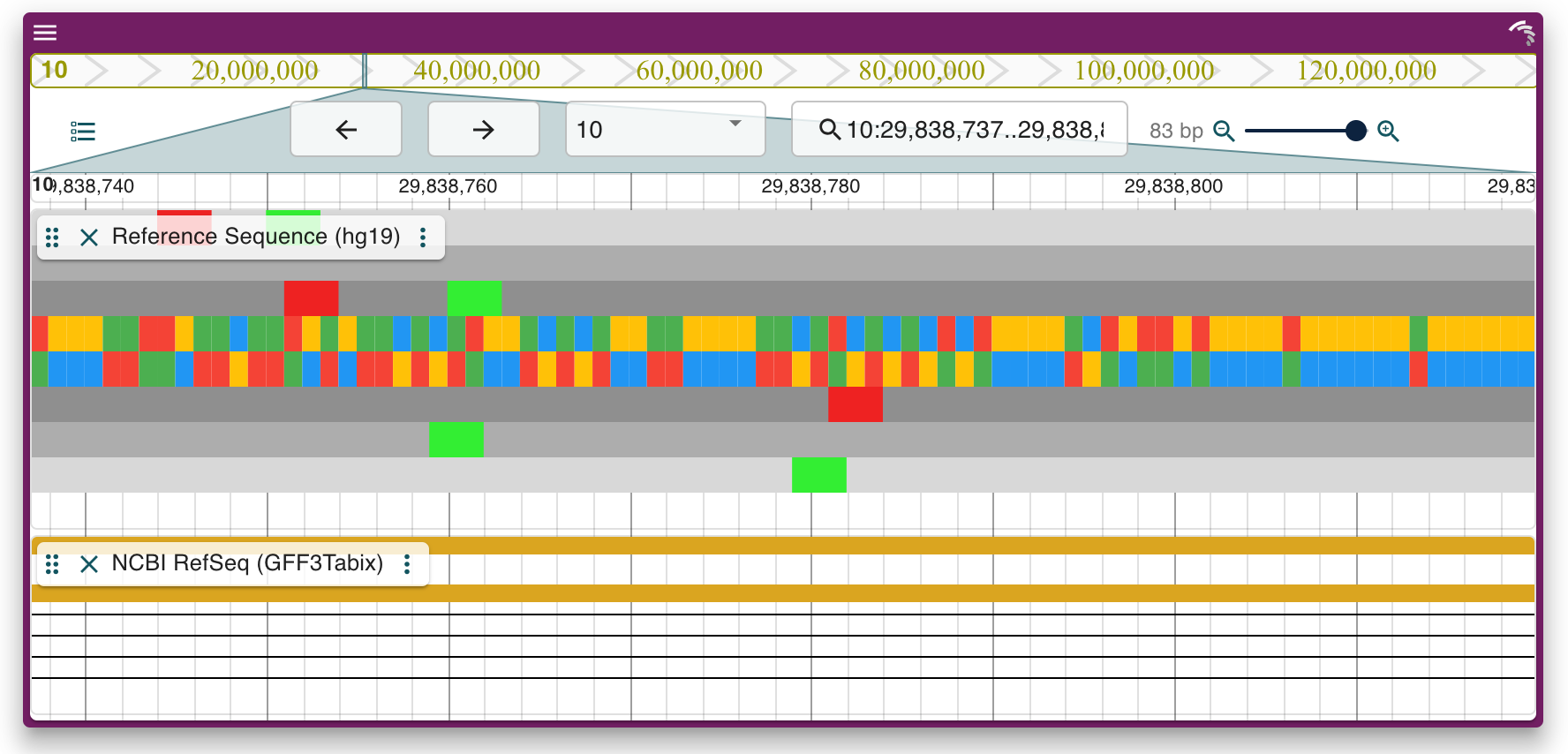
JBrowseR("ViewHg19", location = "10:29,838,737..29,838,819")In order to get started with JBrowseR, please refer to the vignette that best suits your needs:
Allows you to search by gene name
- Link demo: https://gmod.shinyapps.io/basic_usage_with_text_index
- Source code: https://github.com/GMOD/JBrowseR/blob/main/example_apps/basic_usage_with_text_index/app.R
Shows a "bookmark" type feature, loading data from data frame, and buttons to navigate to genes of interest
- Live link: https://gmod.shinyapps.io/bookmarks_demo
- Source code: https://github.com/GMOD/JBrowseR/blob/main/example_apps/bookmarks_demo/app.R
Shows loading a config.json file
- Live link: https://gmod.shinyapps.io/load_config_json
- Source code: https://github.com/GMOD/JBrowseR/blob/main/example_apps/load_config_json/app.R
Simple example showing a data frame as a track
- Live link: https://gmod.shinyapps.io/load_data_frame
- Source code: https://github.com/GMOD/JBrowseR/blob/main/example_apps/load_data_frame/app.R
Shows putative gene fusions in the SKBR3 breast cancer cell line
- Live link: https://gmod.shinyapps.io/skbr3_gene_fusions
- Source code: https://github.com/GMOD/JBrowseR/blob/main/example_apps/skbr3_gene_fusions/app.R
Uses the config.json loading method
- Live link: not yet online
- Source code: https://github.com/GMOD/JBrowseR/blob/main/example_apps/using_plugins/app.R
If you use JBrowseR in your research, please cite the following publication:
Hershberg et al., 2021. JBrowseR: An R Interface to the JBrowse 2 Genome Browser
@article{hershberg2021jbrowser,
title={JBrowseR: An R Interface to the JBrowse 2 Genome Browser},
author={Hershberg, Elliot A and Stevens, Garrett and Diesh, Colin and Xie, Peter and De Jesus Martinez, Teresa and Buels, Robert and Stein, Lincoln and Holmes, Ian},
journal={Bioinformatics}
}