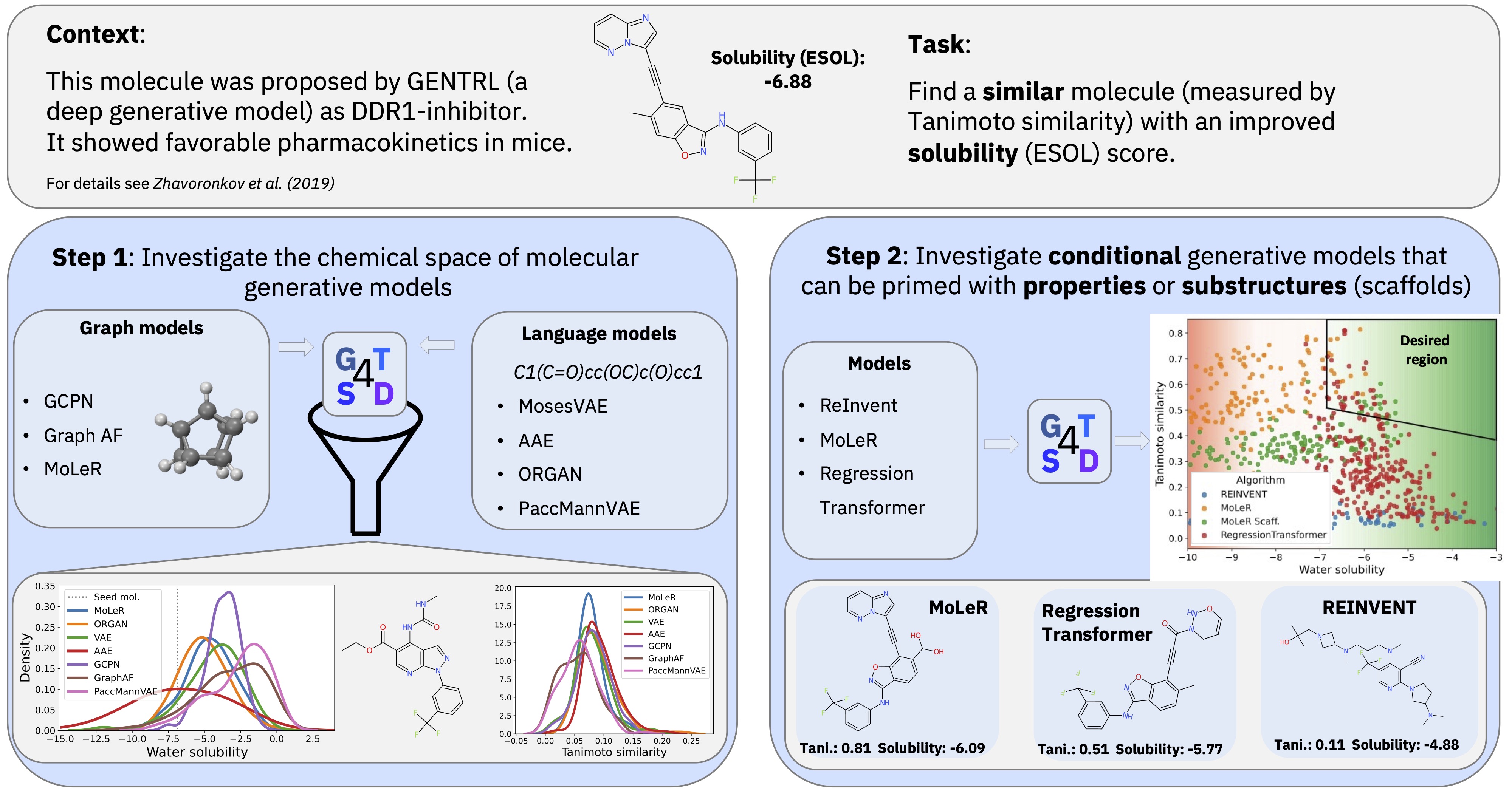
The GT4SD (Generative Toolkit for Scientific Discovery) is an open-source platform to accelerate hypothesis generation in the scientific discovery process. It provides a library for making state-of-the-art generative AI models easier to use.
For full details on the library API and examples see the docs.
Almost all pretrained models are also available via gradio-powered web apps on Hugging Face Spaces.
Currently gt4sd relies on:
- python>=3.7,<=3.10
- pip==24.0
If you need others, help us by contributing to the project.
The recommended way to install the gt4sd is to create a dedicated conda environment, this will ensure all requirements are satisfied. For CPU:
git clone https://github.com/GT4SD/gt4sd-core.git
cd gt4sd-core/
conda env create -f conda_cpu_mac.yml # for linux use conda_cpu_linux.yml
conda activate gt4sd
pip install gt4sdNOTE 1: By default gt4sd is installed with CPU requirements. For GPU usage replace with:
conda env create -f conda_gpu.ymlNOTE 2: In case you want to reuse an existing compatible environment (see requirements), you can use pip, but as of now (:eyes: on issue for changes), some dependencies require installation from GitHub, so for a complete setup install them with:
pip install -r vcs_requirements.txtA few VCS dependencies require Git LFS (make sure it's available on your system).
If you would like to contribute to the package, we recommend to install gt4sd in
editable mode inside your conda environment:
pip install --no-deps -e .Learn more in CONTRIBUTING.md
After install you can use gt4sd right away in your discovery workflows.
Running an algorithm is as easy as typing:
from gt4sd.algorithms.conditional_generation.paccmann_rl.core import (
PaccMannRLProteinBasedGenerator, PaccMannRL
)
target = 'MVLSPADKTNVKAAWGKVGAHAGEYGAEALERMFLSFPTT'
# algorithm configuration with default parameters
configuration = PaccMannRLProteinBasedGenerator()
# instantiate the algorithm for sampling
algorithm = PaccMannRL(configuration=configuration, target=target)
items = list(algorithm.sample(10))
print(items)Or you can use the ApplicationRegistry to run an algorithm instance using a serialized representation of the algorithm:
from gt4sd.algorithms.registry import ApplicationsRegistry
target = 'MVLSPADKTNVKAAWGKVGAHAGEYGAEALERMFLSFPTT'
algorithm = ApplicationsRegistry.get_application_instance(
target=target,
algorithm_type='conditional_generation',
domain='materials',
algorithm_name='PaccMannRL',
algorithm_application='PaccMannRLProteinBasedGenerator',
generated_length=32,
# include additional configuration parameters as **kwargs
)
items = list(algorithm.sample(10))
print(items)GT4SD can run inference pipelines based on the gt4sd-inference CLI command.
It allows to run all inference algorithms directly from the command line.
To see which algorithms are available and how to use the CLI for your favorite model,
check out examples/cli/README.md.
You can run inference pipelines simply typing:
gt4sd-inference --algorithm_name PaccMannRL --algorithm_application PaccMannRLProteinBasedGenerator --target MVLSPADKTNVKAAWGKVGAHAGEYGAEALERMFLSFPTT --number_of_samples 10The command supports multiple parameters to select an algorithm and configure it for inference:
$ gt4sd-inference --help
usage: gt4sd-inference [-h] [--algorithm_type ALGORITHM_TYPE]
[--domain DOMAIN] [--algorithm_name ALGORITHM_NAME]
[--algorithm_application ALGORITHM_APPLICATION]
[--algorithm_version ALGORITHM_VERSION]
[--target TARGET]
[--number_of_samples NUMBER_OF_SAMPLES]
[--configuration_file CONFIGURATION_FILE]
[--print_info [PRINT_INFO]]
optional arguments:
-h, --help show this help message and exit
--algorithm_type ALGORITHM_TYPE
Inference algorithm type, supported types:
conditional_generation, controlled_sampling,
generation, prediction. (default: None)
--domain DOMAIN Domain of the inference algorithm, supported types:
materials, nlp. (default: None)
--algorithm_name ALGORITHM_NAME
Inference algorithm name. (default: None)
--algorithm_application ALGORITHM_APPLICATION
Inference algorithm application. (default: None)
--algorithm_version ALGORITHM_VERSION
Inference algorithm version. (default: None)
--target TARGET Optional target for generation represented as a
string. Defaults to None, it can be also provided in
the configuration_file as an object, but the
commandline takes precendence. (default: None)
--number_of_samples NUMBER_OF_SAMPLES
Number of generated samples, defaults to 5. (default:
5)
--configuration_file CONFIGURATION_FILE
Configuration file for the inference pipeline in JSON
format. (default: None)
--print_info [PRINT_INFO]
Print info for the selected algorithm, preventing
inference run. Defaults to False. (default: False)You can use gt4sd-inference to directly get information on the configuration parameters for the selected algorithm:
gt4sd-inference --algorithm_name PaccMannRL --algorithm_application PaccMannRLProteinBasedGenerator --print_info
INFO:gt4sd.cli.inference:Selected algorithm: {'algorithm_type': 'conditional_generation', 'domain': 'materials', 'algorithm_name': 'PaccMannRL', 'algorithm_application': 'PaccMannRLProteinBasedGenerator', 'algorithm_version': 'v0'}
INFO:gt4sd.cli.inference:Selected algorithm support the following configuration parameters:
{
"batch_size": {
"description": "Batch size used for the generative model sampling.",
"title": "Batch Size",
"default": 32,
"type": "integer",
"optional": true
},
"temperature": {
"description": "Temperature parameter for the softmax sampling in decoding.",
"title": "Temperature",
"default": 1.4,
"type": "number",
"optional": true
},
"generated_length": {
"description": "Maximum length in tokens of the generated molcules (relates to the SMILES length).",
"title": "Generated Length",
"default": 100,
"type": "integer",
"optional": true
}
}
Target information:
{
"target": {
"title": "Target protein sequence",
"description": "AA sequence of the protein target to generate non-toxic ligands against.",
"type": "string"
}
}GT4SD provides a trainer client based on the gt4sd-trainer CLI command.
The trainer currently supports the following training pipelines:
language-modeling-trainer: language modelling via HuggingFace transfomers and PyTorch Lightning.paccmann-vae-trainer: PaccMann VAE models.granular-trainer: multimodal compositional autoencoders supporting MLP, RNN and Transformer layers.guacamol-lstm-trainer: GuacaMol LSTM models.moses-organ-trainer: Moses Organ implementation.moses-vae-trainer: Moses VAE models.torchdrug-gcpn-trainer: TorchDrug Graph Convolutional Policy Network model.torchdrug-graphaf-trainer: TorchDrug autoregressive GraphAF model.diffusion-trainer: Diffusers model.gflownet-trainer: GFlowNet model.
$ gt4sd-trainer --help
usage: gt4sd-trainer [-h] --training_pipeline_name TRAINING_PIPELINE_NAME
[--configuration_file CONFIGURATION_FILE]
optional arguments:
-h, --help show this help message and exit
--training_pipeline_name TRAINING_PIPELINE_NAME
Training type of the converted model, supported types:
granular-trainer, language-modeling-trainer, paccmann-
vae-trainer. (default: None)
--configuration_file CONFIGURATION_FILE
Configuration file for the trainining. It can be used
to completely by-pass pipeline specific arguments.
(default: None)To launch a training you have two options.
You can either specify the training pipeline and the path of a configuration file that contains the needed training parameters:
gt4sd-trainer --training_pipeline_name ${TRAINING_PIPELINE_NAME} --configuration_file ${CONFIGURATION_FILE}Or you can provide directly the needed parameters as arguments:
gt4sd-trainer --training_pipeline_name language-modeling-trainer --type mlm --model_name_or_path mlm --training_file /path/to/train_file.jsonl --validation_file /path/to/valid_file.jsonlTo get more info on a specific training pipeleins argument simply type:
gt4sd-trainer --training_pipeline_name ${TRAINING_PIPELINE_NAME} --helpOnce a training pipeline has been run via the gt4sd-trainer, it's possible to save the trained algorithm via gt4sd-saving for usage in compatible inference pipelines.
Here a small example for PaccMannGP algorithm (paper).
You can train a model with gt4sd-trainer (quick training using few data, not really recommended for a realistic model
gt4sd-trainer --training_pipeline_name paccmann-vae-trainer --epochs 250 --batch_size 4 --n_layers 1 --rnn_cell_size 16 --latent_dim 16 --train_smiles_filepath src/gt4sd/training_pipelines/tests/molecules.smi --test_smiles_filepath src/gt4sd/training_pipelines/tests/molecules.smi --model_path /tmp/gt4sd-paccmann-gp/ --training_name fast-example --eval_interval 15 --save_interval 15 --selfiesSave the model with the compatible inference pipeline using gt4sd-saving:
gt4sd-saving --training_pipeline_name paccmann-vae-trainer --model_path /tmp/gt4sd-paccmann-gp/ --training_name fast-example --target_version fast-example-v0 --algorithm_application PaccMannGPGeneratorRun the algorithm via gt4sd-inference (again the model produced in the example is trained on dummy data and will give dummy outputs, do not use it as is 🙅):
gt4sd-inference --algorithm_name PaccMannGP --algorithm_application PaccMannGPGenerator --algorithm_version fast-example-v0 --number_of_samples 5 --target '{"molwt": {"target": 60.0}}'You can upload trained and finetuned models easily in the public hub using gt4sd-upload. The syntax follows the saving pipeline:
gt4sd-upload --training_pipeline_name paccmann-vae-trainer --model_path /tmp/gt4sd-paccmann-gp --training_name fast-example --target_version fast-example-v0 --algorithm_application PaccMannGPGeneratorNOTE: GT4SD can be configured to upload models to a custom or self-hosted COS. An example on self-hosting locally a COS (minio) where to upload your models can be found here.
You can compute properties of your generated samples using the gt4sd.properties submodule:
>>>from gt4sd.properties import PropertyPredictorRegistry
>>>similarity_predictor = PropertyPredictorRegistry.get_property_predictor("similarity_seed", {"smiles" : "C1=CC(=CC(=C1)Br)CN"})
>>>similarity_predictor("CCO")
0.0333
>>># let's inspect what other parameters we can set for similarity measuring
>>>similarity_predictor = PropertyPredictorRegistry.get_property_predictor("similarity_seed", {"smiles" : "C1=CC(=CC(=C1)Br)CN", "fp_key": "ECFP6"})
>>>similarity_predictor("CCO")
>>># inspect parameters
>>>PropertyPredictorRegistry.get_property_predictor_parameters_schema("similarity_seed")
'{"title": "SimilaritySeedParameters", "description": "Abstract class for property computation.", "type": "object", "properties": {"smiles": {"title": "Smiles", "example": "c1ccccc1", "type": "string"}, "fp_key": {"title": "Fp Key", "default": "ECFP4", "type": "string"}}, "required": ["smiles"]}'
>>># predict other properties
>>>qed = PropertyPredictorRegistry.get_property_predictor("qed")
>>>qed('CCO')
0.4068
>>># list properties
>>>PropertyPredictorRegistry.list_available()
['activity_against_target',
'aliphaticity',
...
'scscore',
'similarity_seed',
'tpsa',
'weight']Find more examples in notebooks
You can play with them right away using the provided Dockerfile, simply build the image and run it to explore the examples using Jupyter:
docker build -f Dockerfile -t gt4sd-demo .
docker run -p 8888:8888 gt4sd-demoBeyond implementing various generative modeling inference and training pipelines GT4SD is designed to provide a high-level API that implement an harmonized interface for several existing packages:
- GuacaMol: inference pipelines for the baselines models and training pipelines for LSTM models.
- Moses: inference pipelines for the baselines models and training pipelines for VAEs and Organ.
- TorchDrug: inference and training pipelines for GCPN and GraphAF models. Training pipelines support custom datasets as well as datasets native in TorchDrug.
- MoLeR: inference pipelines for MoLeR (MOlecule-LEvel Representation) generative models for de-novo and scaffold-based generation.
- TAPE: encoder modules compatible with the protein language models.
- PaccMann: inference pipelines for all algorithms of the PaccMann family as well as training pipelines for the generative VAEs.
- transformers: training and inference pipelines for generative models from HuggingFace Models
- diffusers: training and inference pipelines for generative models from Diffusers Models
- GFlowNets: training and inference pipeline for Generative Flow Networks
- MolGX: training and inference pipelines to generate small molecules satisfying target properties. The full implementation of MolGX, including additional functionalities, is available here.
- Regression Transformers: training and inference pipelines to generate small molecules, polymers or peptides based on numerical property constraints. For details read the paper.
If you use gt4sd in your projects, please consider citing the following:
@software{GT4SD,
author = {GT4SD Team},
month = {2},
title = {{GT4SD (Generative Toolkit for Scientific Discovery)}},
url = {https://github.com/GT4SD/gt4sd-core},
version = {main},
year = {2022}
}
@article{manica2022gt4sd,
title={Accelerating material design with the generative toolkit for scientific discovery},
author={Manica, Matteo and Born, Jannis and Cadow, Joris and Christofidellis, Dimitrios and Dave, Ashish and Clarke, Dean and Teukam, Yves Gaetan Nana and Giannone, Giorgio and Hoffman, Samuel C and Buchan, Matthew and others},
journal={npj Computational Materials},
volume={9},
number={1},
pages={69},
year={2023},
publisher={Nature Publishing Group UK London}
}The gt4sd codebase is under MIT license.
For individual model usage, please refer to the model licenses found in the original packages.