Scalable differential expression analysis of single cell transcriptomics datasets with complex study designs
The dreamlet package enables differential expression analysis on multi-sample single cell datasets using linear (mixed) models with precision weights.
Major functionality of dreamlet package using the Bioconductor SingleCellExperiment interface:
aggregateToPseudoBulk()Fast evaluation of pseudobulk from raw or estimated countsprocessAssays()Normalize aggregated counts, compute precision weightsfitVarPart()Variance partitioning analysisdreamlet()Differential expression analysis across samplesdreamletCompareClusters()Differential expression analysis across cell clusterszenith_gsa()Gene set analysis with full spectrum of test statisticscompositePosteriorTest()Test cell type specifity of effects with Bayesian meta-analysismeta_analysis()Frequentist meta-analysis across cohortsoutlierByAssay()Outlier detection based on gene expression
- Preprint on biorxiv
- Full reproducible analysis code for 4 large-scale datasets
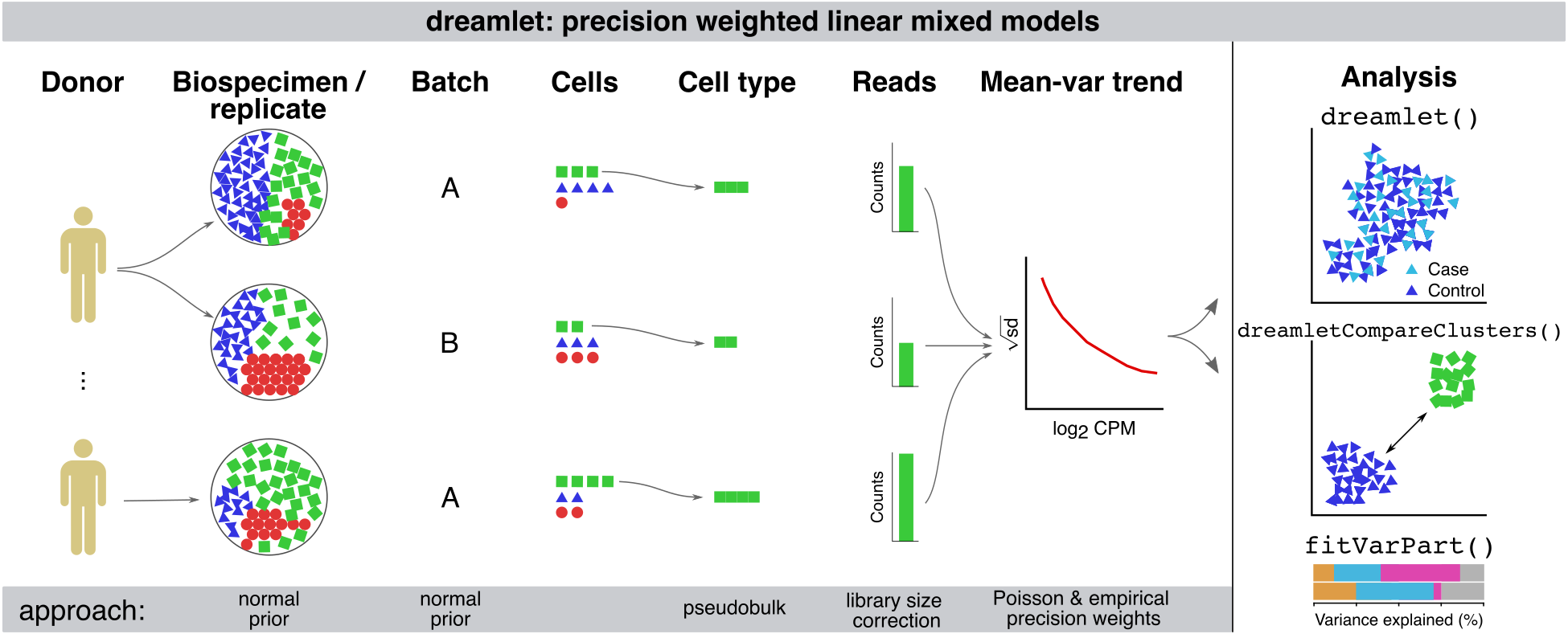
Dreamlet further enables analysis of massive-scale of single cell/nucleus transcriptome datasets by addressing both CPU and memory usage limitations. Dreamlet performs preprocessing and statistical analysis in parallel on multicore machines, and can distribute work across multiple nodes on a compute cluster. Dreamlet also uses the H5AD format for on-disk data storage to enable data processing in smaller chunks to dramatically reduce memory usage.
The dreamlet workflow easily integrates into the Bioconductor ecosystem, and uses the SingleCellExperiment class to facilitate compatibility with other analyses. Beyond differential expression testing, dreamlet provides seamless integration of downstream analysis including quantifying sources of expression variation, gene set analysis using the full spectrum of gene-level t-statistics, testing differences in cell type composition and visualizing results.
Dreamlet builds on previous work on variance partitioning and differential expression using precision-weighted linear mixed models in the variancePartition package. The dreamlet package is designed to be easily adopted by users of variancePartition and limma.
dreamlet >= 1.0.0 is compatible with BioC v3.18 for R v4.3.
# 1) Make sure Bioconductor is installed
# 2) Install dreamlet and dependencies:
devtools::install_github("DiseaseNeurogenomics/dreamlet")
# 3) Install zellkonverter >= v1.10.1
BiocManager::install("zellkonverter")dreamlet is also compatible with earlier version of R and Bioconductor after installing these dependencies:
In case these aren't installed automatically:
devtools::install_github("DiseaseNeurogenomics/variancePartition")
devtools::install_github("DiseaseNeurogenomics/zenith")