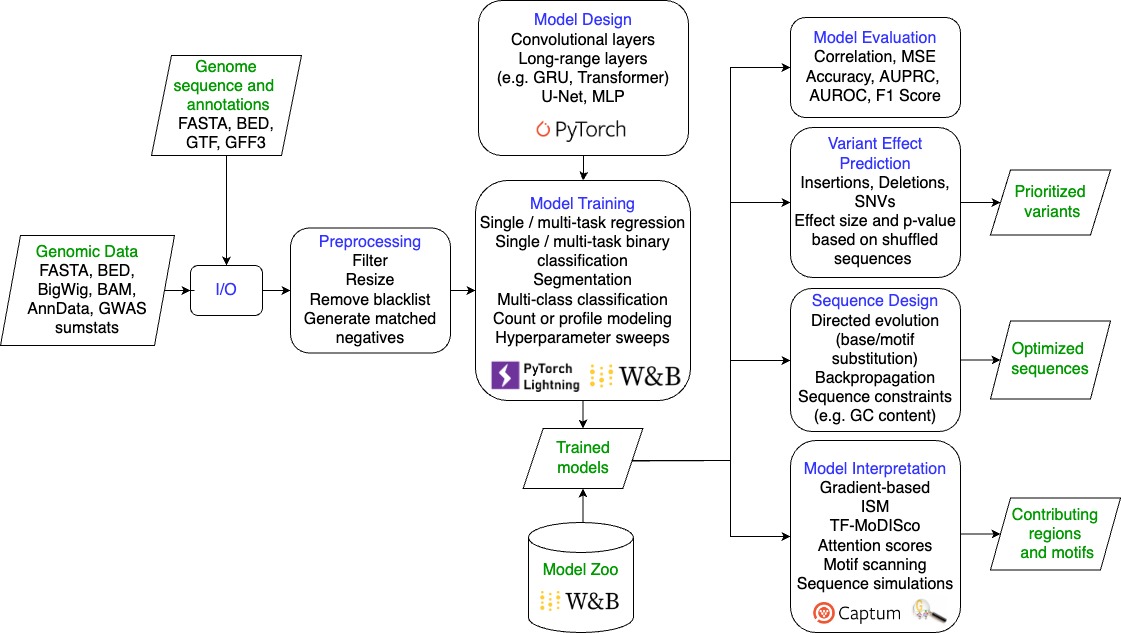
gReLU is a Python library to train, interpret, and apply deep learning models to DNA sequences. Code documentation is available here.
To install from source:
git clone https://github.com/Genentech/gReLU.git
cd gReLU
pip install .To install using pip:
pip install gReLUThis project uses pre-commit. Please make sure to install it before making any changes:
pip install pre-commit
cd gReLU
pre-commit installIt is a good idea to update the hooks to the latest version:
pre-commit autoupdateIf you want to use genome annotation features through the function grelu.io.genome.read_gtf, you will need to install the following UCSC utilities: genePredToBed, genePredToGtf, bedToGenePred, gtfToGenePred, gff3ToGenePred.
If you want to create bigWig files through the function grelu.data.preprocess.make_insertion_bigwig, you will need to install the following UCSC utilities: bedGraphToBigWig.
UCSC utilities can be installed from http://hgdownload.cse.ucsc.edu/admin/exe/, for example using the following commands:
rsync -aP rsync://hgdownload.soe.ucsc.edu/genome/admin/exe/linux.x86_64/bedGraphToBigWig /usr/bin/
rsync -aP rsync://hgdownload.soe.ucsc.edu/genome/admin/exe/linux.x86_64/genePredToBed /usr/bin/
rsync -aP rsync://hgdownload.soe.ucsc.edu/genome/admin/exe/linux.x86_64/genePredToGtf /usr/bin/
rsync -aP rsync://hgdownload.soe.ucsc.edu/genome/admin/exe/linux.x86_64/bedToGenePred /usr/bin/
rsync -aP rsync://hgdownload.soe.ucsc.edu/genome/admin/exe/linux.x86_64/gtfToGenePred /usr/bin/
rsync -aP rsync://hgdownload.soe.ucsc.edu/genome/admin/exe/linux.x86_64/gff3ToGenePred /usr/bin/or via bioconda:
conda install -y \
bioconda::ucsc-bedgraphtobigwig \
bioconda::ucsc-genepredtobed \
bioconda::ucsc-genepredtogtf \
bioconda::ucsc-bedtogenepred \
bioconda::ucsc-gtftogenepred \
bioconda::ucsc-gff3togenepred