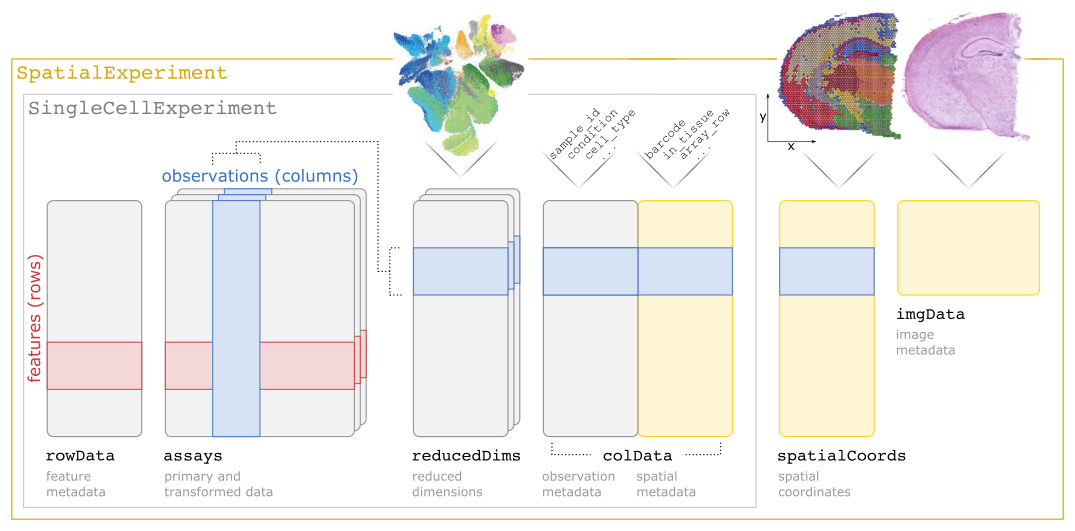
SpatialExperiment is an R/Bioconductor S4 class for storing data from spatially resolved transcriptomics (ST) experiments. The class extends the SingleCellExperiment class for single-cell data to support storage and retrieval of additional information from spot-based and molecule-based ST platforms, including spatial coordinates, images, and image metadata. A specialized constructor function is included for data from the 10x Genomics Visium platform.
The SpatialExperiment package is available from Bioconductor.
A vignette containing examples and documentation is available from Bioconductor, and additional details are provided in our preprint.
The following schematic illustrates the SpatialExperiment class structure.
The SpatialExperiment package can be installed from Bioconductor. Note that Bioconductor follows a "release" and "development" schedule, where the release version is considered to be stable and updated every 6 months, and the development version contains latest updates (and then becomes the next release version every 6 months).
To install the stable release version, install the latest release version of R from CRAN, then install the Bioconductor package installer and the SpatialExperiment package as follows.
install.packages("BiocManager")
BiocManager::install("SpatialExperiment")
To install the development version, there are two options.
(i) Install the development version of the SpatialExperiment package from GitHub, using a standard installation of the release version of R from CRAN:
install.packages("remotes")
remotes::install_github("drighelli/SpatialExperiment")
(ii) Install a complete devel version of Bioconductor. This requires first installing the appropriate version of R (currently R-devel). Then, install the Bioconductor package installer and the development version of the SpatialExperiment package:
install.packages("BiocManager")
BiocManager::install("SpatialExperiment", version = "devel")
In version 1.5.2 the spatialData slot was deprecated and columns previously stored in spatialData moved to colData, and in version 1.5.3 the internal class SpatialImage was renamed to VirtualSpatialImage to avoid a clash with the same class name in Seurat. We recommend updating to version 1.5.3 (or later) and rebuilding existing objects to ensure longer-term compatibility with future versions.
To use version 1.5.3 or later (as of March 2022), please follow the installation instructions for the development version above.
(These updates will also be available in the release version from mid-April 2022 onwards, following the Bioconductor release schedule.)
Righell D.*, Weber L.M.*, Crowell H.L.*, Pardo B., Collado-Torres L., Ghazanfar S., Lun A.T.L., Hicks S.C.+, and Risso D.+ (2021), SpatialExperiment: infrastructure for spatially resolved transcriptomics data in R using Bioconductor, bioRxiv.