This is the package of our winning algorithm in the 2018 "You Snooze, You Win" PhysioNet Challenge. Details can be found from Hongyang Li, Yuanfang Guan. DeepSleep convolutional neural network allows accurate and fast detection of sleep arousal. Communications Biology 2021.. Please contact (hyangl@umich.edu or gyuanfan@umich.edu) if you have any questions or suggestions.
background: 2018 PhysioNet Challenge
Git clone a copy of code:
git clone https://github.com/GuanLab/DeepSleep.git
- python (3.6.5)
- numpy (1.13.3). It comes pre-packaged in Anaconda.
- scikit-learn (0.19.0) A popular machine learning package. It can be installed by:
pip install -U scikit-learn
- tensorflow (1.8.0)
- keras (2.0.4)
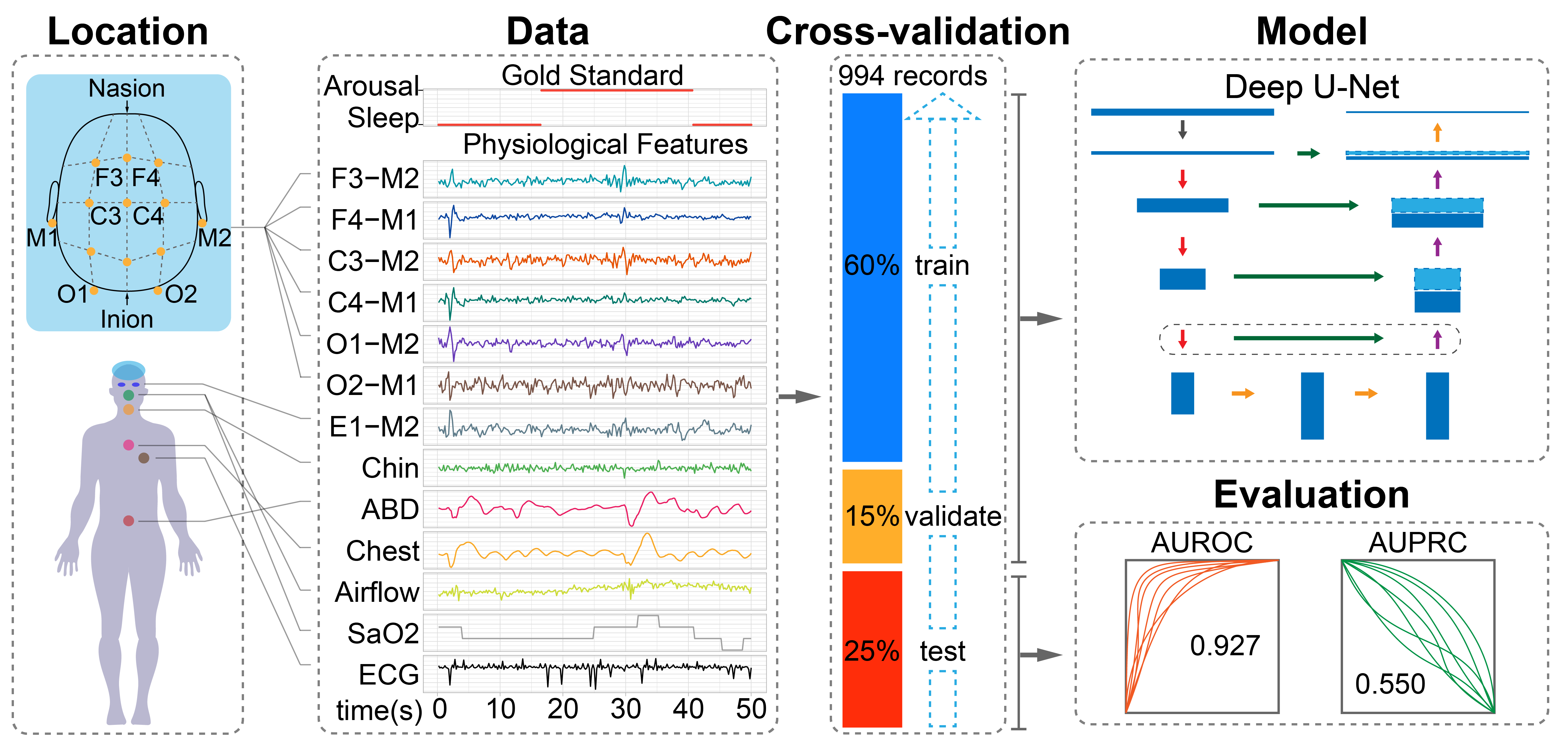
We used the public training data from the challenge, which contains 994 polysomnographic recordings. Each polysomnogram has 13 channels, corresponding to 6 EEG (F3-M2, F4-M1, C3-M2, C4-M1, O1-M2, O2-M1), 3 EMG (chin, abdominal, chest), Airflow, Saturation of oxygen, and ECG. These polysomnogram have been manually labeled by sleep experts, including sleep stages, arousals, apneas and other sleep events. Download link and details can be found HERE.
First download the data and put them into the folder "./data/training/". The training dataset is approximately 135 GB in size. Or you can try our code using the examples provided but unzip them first:
cd ./data/training/
unzip tr03-0078.zip
unzip tr03-0079.zip
cd ../../
Since the lengths of sleep recordings are different, we first make uniform these recordings to the same 8-million length (2^23 = 8,388,608) by padding zeros at both the beginning and the end.
unzip ref555.zip
python uniform_8m.py
Now you can run predictions by providing the sample id:
python predict.py tr03-0078
It will generate a file called "tr03-0078.vec", each line corresponds to the prediction for each time point in the original polysomnogram.
If you want to run multiple predictions, you can try:
python predict.py tr03-0078 tr03-0079
The AUPRC and AUROC can be calculated by running:
python score2018.py *vec -r ./data/training/
It will print out the results like this:
record AUROC AUPRC
_______________________________
tr03-0078 0.932568 0.554207
tr03-0079 0.975179 0.314851
_______________________________
Overall 0.950086 0.492553
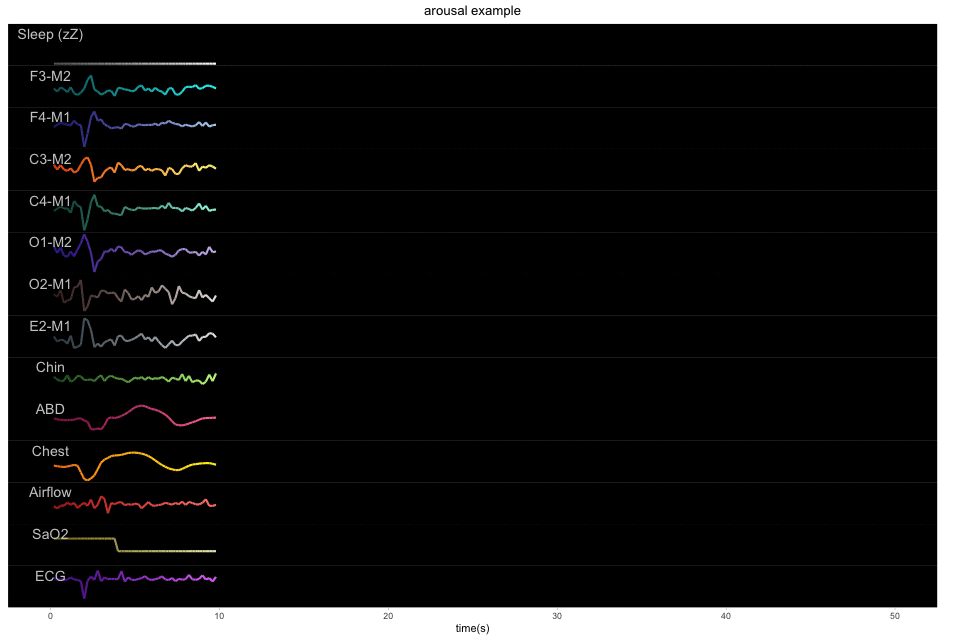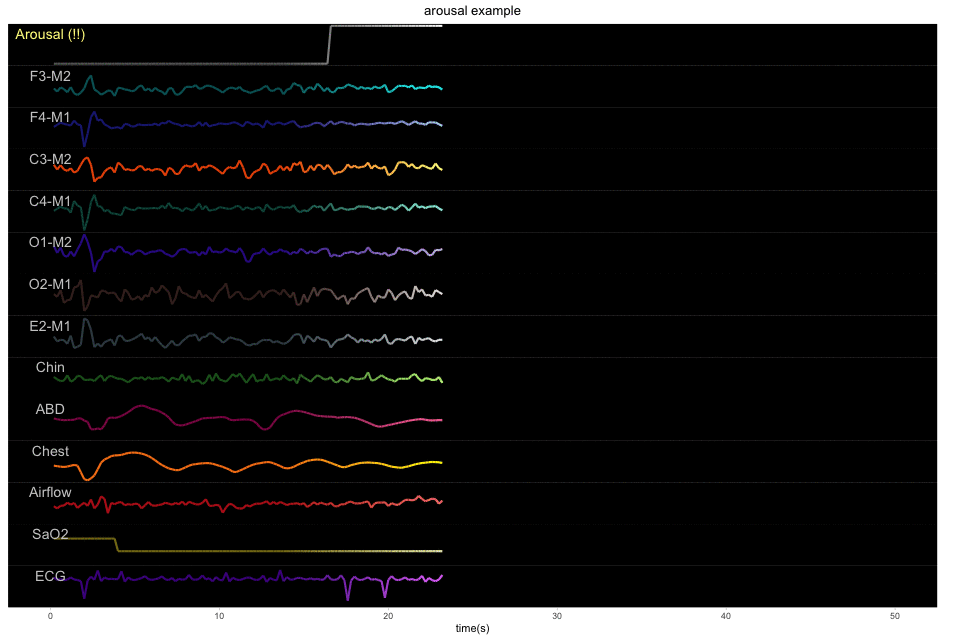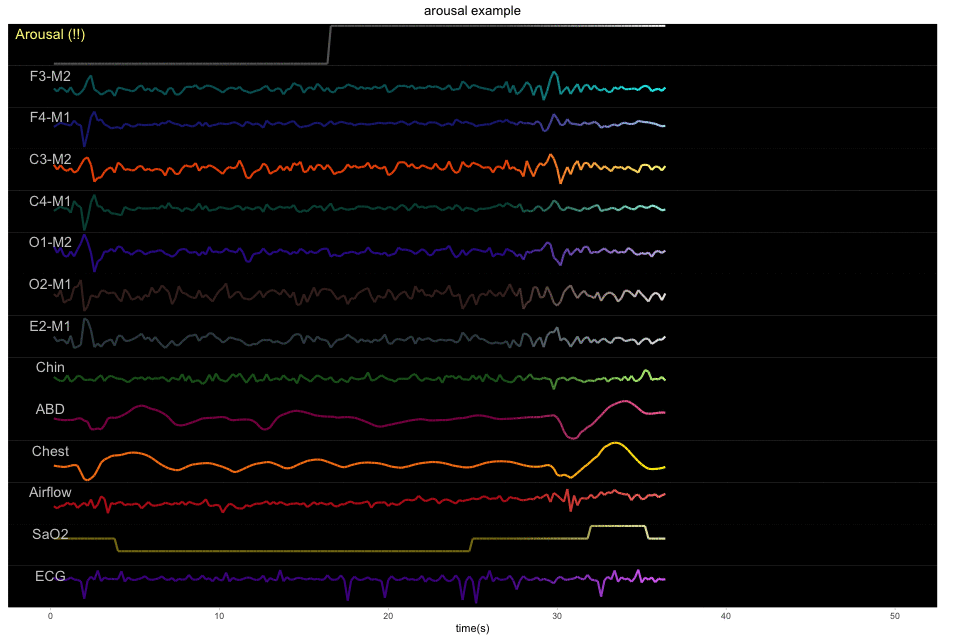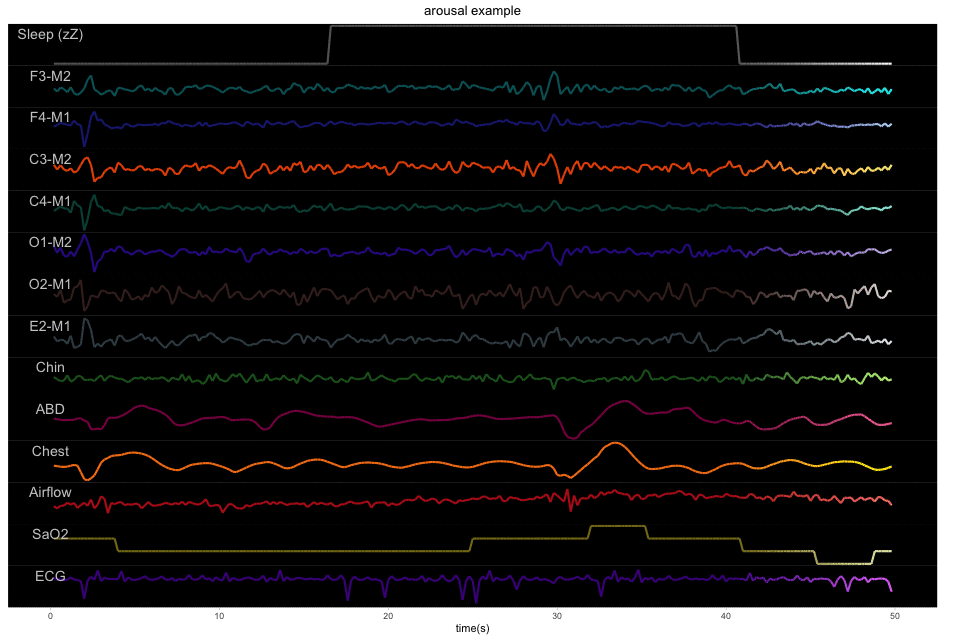
A 50-second example with 13 physiological signals and the corresponding binary label of sleep or arousal:
Here the two example records were used to train the model. In practice, please download the PhysioNet Challenge dataset or your own dataset to train the model. In the example "train.py" and "predict.py", we used five input channels:
-
- randomly selected one EEG/EOG channel
-
- randomly selected one EMG channel from Chest or ABD
-
- EMG - Chin
-
- Airflow
-
- ECG You can change the input channels to train models based on your need.
python train.py
We designed three neural networks accept different lengths of input at full, 1/2 and 1/8 resolutions, corresponding to "unet0.py", "unet1.py", and "unet2.py". To run the two lower 1/2 and 1/8 resolution models, the train and predict codes need to be modified.