Authors: Mengyuan Kan, Avantika Diwadkar, Maya Shumyatcher, Blanca Himes
RAVED (Reproducible Analysis and Validation of Expression Data) is a pipeline that adopts several existing informatics tools but facilitates analysis of publicly available microarray and RNA-Seq data from GEO or SRA. RAVED consists of R Markdown documents that can be modified to execute analysis steps and create summary reports and result files. Here are major steps followed by RAVED:
- GEO/SRA Data Download
- Phenotype Preparation
- Read Alignments and/or Transcript Quantification (RNA-Seq data only)
- Quality Control Metrics
- Differential Expression (DE) Analysis
Several freely accessible R packages for microarray data analysis are used in this pipeline. We use GEO dataset GSE4917 as an example and provide two R Markdown templates to reproduce the analyses and two .html summary reports. The template_QC_report.Rmd script goes first including the steps of GEO data download, phenotype file preparation, and quality metrics measurement. The template_DE_analysis_report.Rmd takes the phenotype file obtained from the template_QC_report.Rmd and performs the differential expression analysis. The RMD files for other microarray data used for the integration analysis of asthma transcriptomic data are available on GitHub Asthma_Transcriptomic_Analysis.
Prerequisite R packages for microarray data analysis include:
- Data download: GEOquery
- Raw data processing: oligo and affy
- Statistical analysis for outlier detection: Hmisc
- DE analysis: limma
- Batch effect adjustment: sva
- Microarry data gene annotation: annotation, hgu133plus2.db and hugene10sttranscriptcluster.db
Raw data are downloaded from GEO directly including GSEMatrix files that are used for phenotype file preparation, and raw intensity data .CEL files, if provided, that are used for QC and DE analysis.
Raw phenotype data derived from GSEMatrix files is first overviewed and then manually modified to a more informative and concise phenotype file with the columns GEO_ID, Subject, Disease, Treatment, Age, Gender, etc
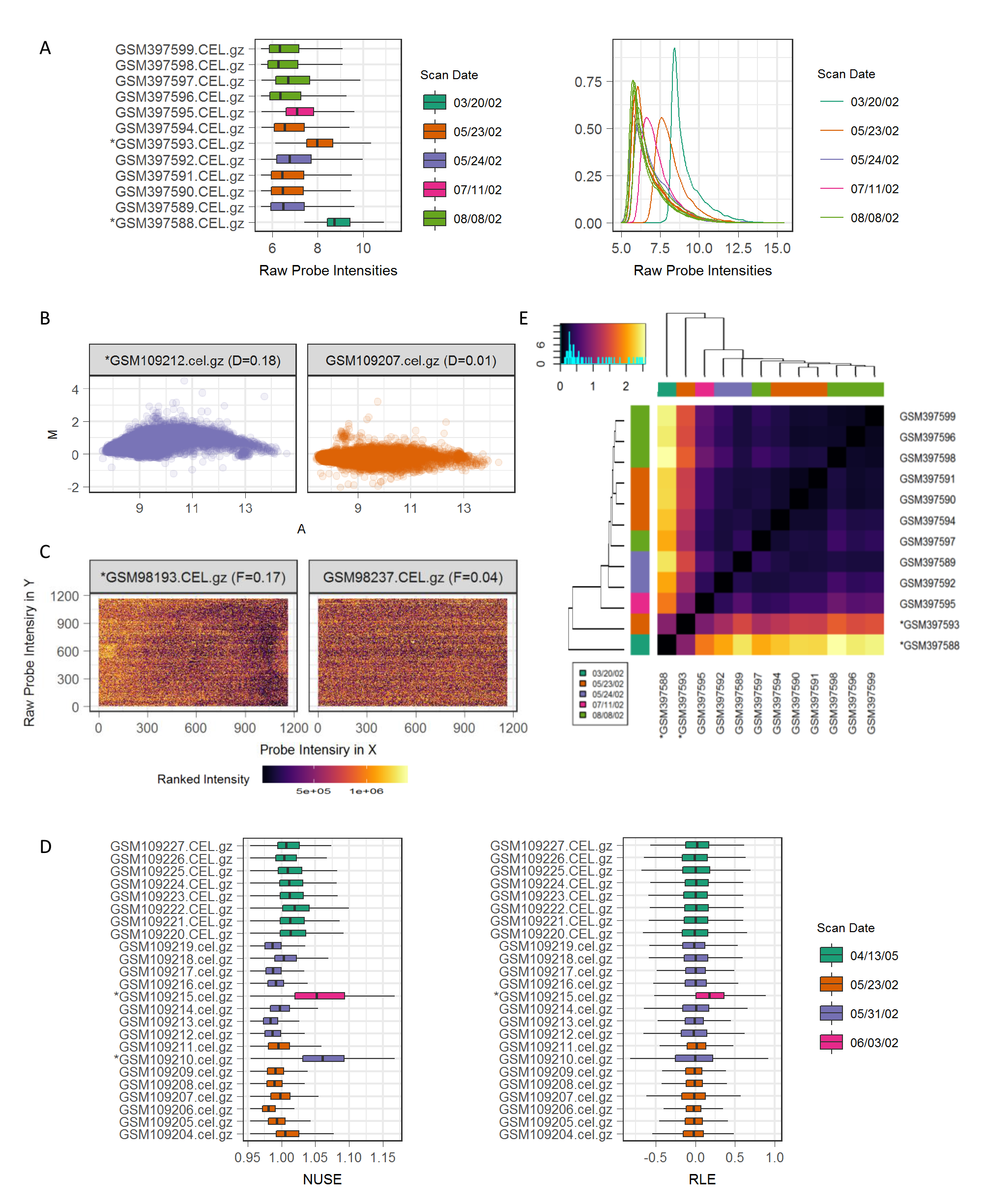
Major QC structure and scoring methods for outliers were adapted from arrayQualityMetrics. The threshold to determine an outlier used in arrayQualityMetrics is the boxplot’s upper whisker, i.e. values beyond 1.5 times the interquartile range, which is also applied to our pipeline. The following QC metrics are included in a routine analysis. The QC metrics used for outlier detection are marked with an asterisk.
- Boxplots and density plots for raw probe intensities*
- RNA degradation plots
- Density plots for perfect match (PM) and mismatch (MM) probe
- MA plots*
- Spatial plots*
- Boxplots for the normalized unscaled standard error (NUSE)*
- Boxplots for the relative log expression (RLE)*
- Heatmap and dendrogram for distance between arrays*
- Principal component analysis (PCA) plots
Raw probe intensities are log2 transformed and visualized by boxplots and density curves. Outlier detection is applied by computing a Kolmogorov-Smirnov statistic (Ka) between log-intensity distribution for one array and the pooled array data, where an array with a Ka beyond the upper whisker is designated as an outlier (Figure 1A).
RNA degradation plots from R package affy assess overall RNA quality, plotting the 5’ to 3’ end of average intensity of each probe base across all probe sets. A high slope from 5’ to 3’ indicates degraded RNA.
Perfect match (PM) and mismatch (MM) probe intensities are log2 transformed and visualized using density curves, where MM probes are expected to have smaller log-intensity at the peak than PM probes due to their nonspecific hybridization.
MA plots compare the log-intensity of a probe set to a median log-intensity of the corresponding probe set across arrays (i.e. reference), measuring the systematic bias in probe levels in an individual array, where the y-axis represents the difference between log-intensity of one probe set to the reference (M or minus), and the x-axis represents the average log-intensity of one probe set and the reference (A or add) (Figure 1B). The MA-plot is expected to be centered on the y-axis M=0 if no significant difference is observed in probe levels across arrays. Outlier detection is applied by computing a Hoeffding's statistic (Da) on the joint distribution of A and M for each array, where an array with a Da >0.15 is designated as an outlier (Figure 1B).
Spatial plots show an artificial colored image of an array’s spatial distribution of intensities that indicate spatial variation in an array. Log-intensities of probes are plotted by their corresponding spatial x and y-coordinate in the array and are expected to be uniformly distributed if the array data has good quality. Outlier detection is applied by computing a sum of the absolute values of low frequency Fourier coefficients (Fa) across all probe sets for each array, where an array with a Fa beyond the upper whisker is designated as an outlier (Figure 1C).
The normalized unscaled standard error (NUSE) and relative log expression (RLE) boxplots indicate probe set homogeneity in one array (Figure 1D), where the metrics are derived from a fitted probe level model by the fitProbeLevelModel function (oligo). The NUSE plots show the distribution of normalized standard error estimates, expected to be centered near 1. Outlier detection is applied by computing an upper hinge (Na) across all probe sets for each array, where an array with a Na beyond the upper whisker is designated as an outlier. The RLE plots represent the distribution of the ratio between the log-intensity of a probe set and the median log-intensity of the corresponding probe set across all arrays, expected to be centered near 0, as a log scale is applied. Outlier detection is applied by computing a Kolmogorov-Smirnov statistic (Ra) between RLE distribution for one array and the pooled array data, where an array with a Ra beyond the upper whisker is designated as an outlier (Figure 1D).
Distance between arrays is evaluated using mean absolute difference of log-intensity between each pair of arrays, where the hierarchical tree between arrays is created based on the distance, which is visualized by a heatmap and dendrogram (Figure 1E). Outlier detection is applied by computing the sum of the distances of one array to all other arrays (Sa), where an array with a Sa beyond the upper whisker is designated as an outlier (Figure 1E).
Principal component analysis (PCA) is performed using log-intensities of all probe sets. PCA plots are generated using first two principle components colored by known factors (e.g. treatment conditions, donners and scan dates), visualizing similarities between arrays and these similarities’ correlation to batch effects.
Figure 1. Examples of Quality Control with Outlier Detection. (A) Boxplots and density plots of raw probe log-intensities for arrays in GSE15823. (B) MA plots for two arrays with smallest and largest scoring in GSE4917. (C) Spatial plots for two arrays with smallest and largest scoring in GSE4302. D) Boxplots and density plots of NUSE and RLE for arrays in GSE4917. E) Heatmap and dendrogram of array pair-wise distance in GSE15823. Outliers are marked with an asterisk. Scan date indicates batch information.
A hard filter is applied to determine the outliers to remove: a sample which is designated an outlier by more than two methods will be excluded from DE analysis. A column showing whether the sample passes QC is added to the phenotype file. We suggest researchers manually inspect the QC metrics and plots to identify outliers.
The comparison between test condition of interest versus within-study control (e.g. asthma vs. non-asthma samples, or glucocorticoid treatment vs. baseline) is manually assigned for DE analysis. DE analysis is performed in a single comparison condition including:
- Robust multi-array average (RMA) of raw intensity signals
- DE analysis by fitting a linear model and empirical Bayes smoothing
- Batch effect adjustment
Official gene symbols are annotated to DE results. Plots are automatically generated for result visualization.
Prerequisite tools for RNA-Seq data analysis include:
- Data download: SRA Toolkit
- Read Alignment: STAR/HTSeq or kallisto
- Quality Control Metrics: FastQC, SAMtools, Bamtools, Picard Tools
- DE analysis: DESeq2 or sleuth
- Gene annotation: BioMart
The RNA-Seq data analysis pipeline is available on GitHub taffeta.
template_QC_report.Rmd: R markdown codes for GSE4917 QC analysis
template_QC_report.html: HTML report for GSE4917 QC analysis
template_DE_analysis_report.Rmd: R markdown codes for GSE4917 DE analysis
template_DE_analysis_report.html: HTML report for GSE4917 DE analysis
c2.cp.kegg.v7.4.symbols.gmt: KEGG pathway file. Place the file under specified data directory.
c2.cp.reactome.v7.4.symbols.gmt: Reactome pathway file. Place the file under specified data directory.