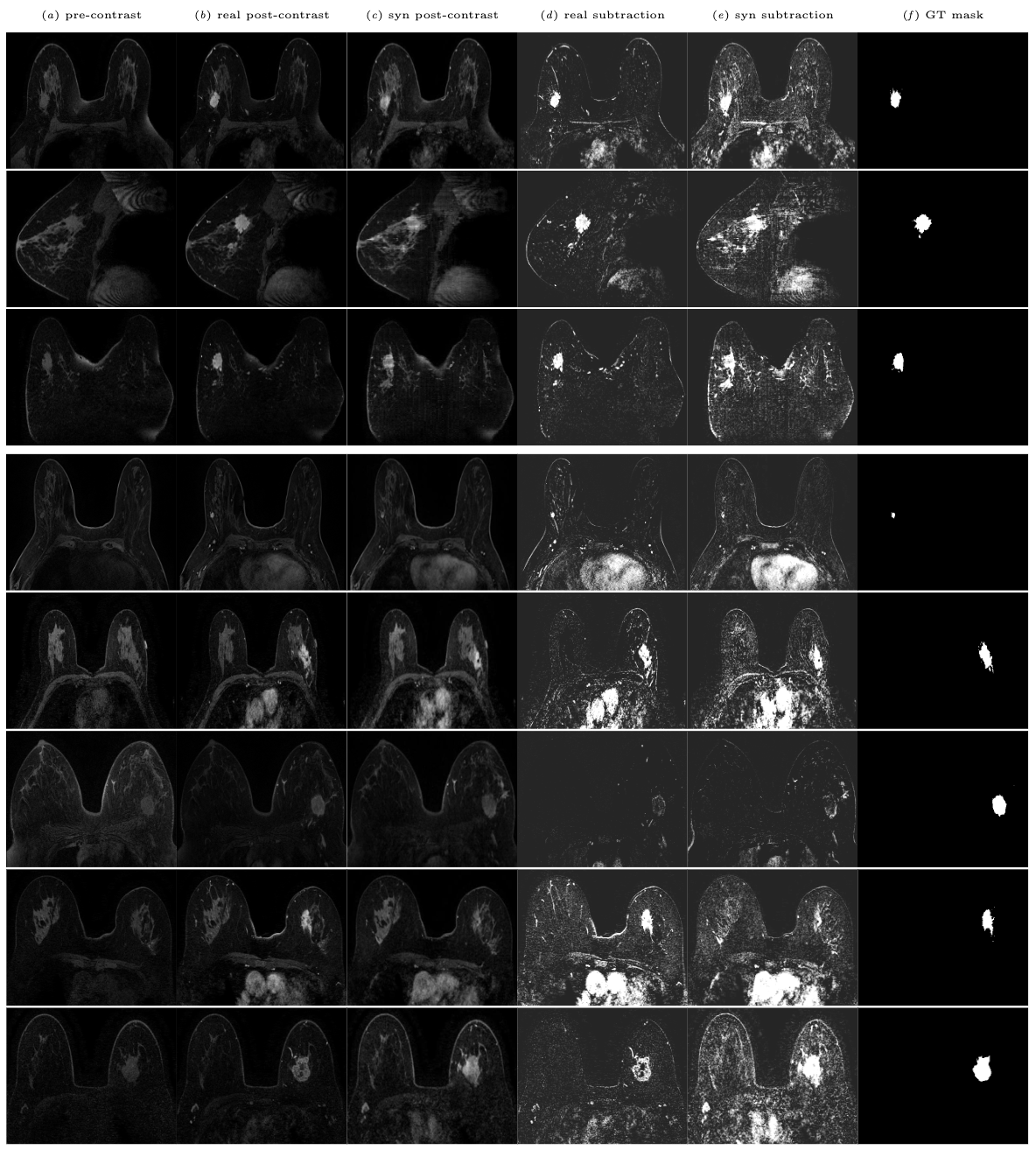
In SPIE Medical Imaging 2024.
The Duke Dataset used in this study is available on The Cancer Imaging Archive (TCIA).
You may find some examples of synthetic nifti files in synthesis/examples.
- Config to run a training of the image synthesis model.
- Config to run a test of the image synthesis model.
- Code to transform Duke DICOM files to NiFti files.
- Code to extract 2D pngs from 3D NiFti files.
- Code to create 3D NiFti files from axial 2D pngs.
- Code to separate synthesis training and test cases.
- Code to compute the image quality metrics such as SSIM, MSE, LPIPS, and more.
- Code to compute the Frèchet Inception Distance (FID) based on ImageNet and RadImageNet.
- Code to prepare 3D single breast cases for nnunet segmentation.
- Train-test-splits of the segmentation dataset.
- Script to run the full nnunet pipeline on the Duke dataset.
Model weights are stored on on Zenodo and made available via the medigan library.
To create your own post-contrast data, simply run:
pip install medigan# import medigan and initialize Generators
from medigan import Generators
generators = Generators()
# generate 10 samples with model 23 (00023_PIX2PIXHD_BREAST_DCEMRI).
# Also, auto-install required model dependencies.
generators.generate(model_id='00023_PIX2PIXHD_BREAST_DCEMRI', num_samples=10, install_dependencies=True)Please consider citing our work if you found it useful for your research:
@article{osuala2023pre,
title={{Pre-to Post-Contrast Breast MRI Synthesis for Enhanced Tumour Segmentation}},
author={Osuala, Richard and Joshi, Smriti and Tsirikoglou, Apostolia and Garrucho, Lidia and Pinaya, Walter HL and Diaz, Oliver and Lekadir, Karim},
journal={arXiv preprint arXiv:2311.10879},
year={2023}
}This repository borrows code from the pix2pixHD and the nnUNet repositories. The 254 tumour segmentation masks used in this study were provided by Caballo et al.