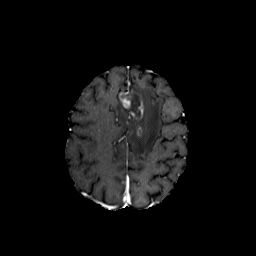
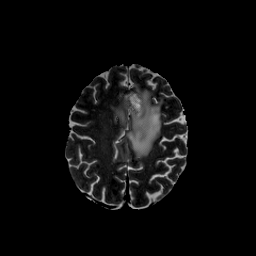
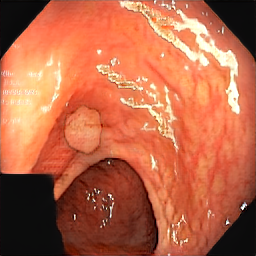
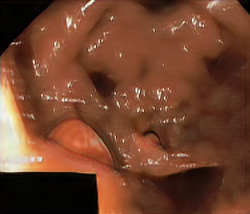
medigan stands for medical generative (adversarial) networks. medigan provides user-friendly medical image synthesis and allows users to choose from a range of pretrained generative models to generate synthetic datasets. These synthetic datasets can be used to train or adapt AI models that perform clinical tasks such as lesion classification, segmentation or detection.
See below how medigan can be run from the command line to generate synthetic medical images.
-
❌ Problem 1: Data scarcity in medical imaging.
-
❌ Problem 2: Scarcity of readily reusable generative models in medical imaging.
-
✅ Solution:
medigan- dataset sharing via generative models 🎁
- data augmentation 🎁
- domain adaptation 🎁
- synthetic data evaluation method testing with multi-model datasets 🎁
Instead of training your own, use one of the generative models from medigan to generate synthetic data.
Search and find a model in medigan using search terms (e.g. "Mammography" or "Endoscopy").
Contribute your own generative model to medigan to increase its visibility, re-use, and impact.
Model information can be found in:
- model documentation (e.g. the parameters of the models' generate functions)
- global.json file (e.g. metadata for model description, selection, and execution)
- medigan paper (e.g. analysis and comparisons of models and FID scores)
To install the current release, simply run:
pip install mediganOr, alternatively via conda:
conda install -c conda-forge mediganExamples and notebooks are located at examples folder
Documentation is available at medigan.readthedocs.io
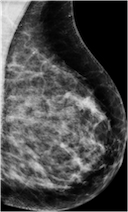
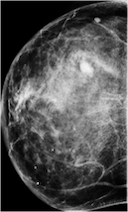
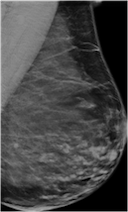
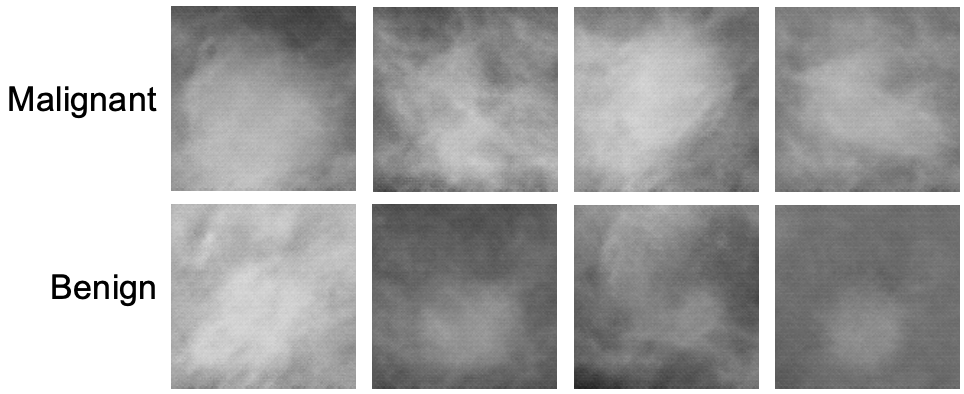
Create mammography masses with labels (malignant or benign) using a class-conditional DCGAN model.
# import medigan and initialize Generators
from medigan import Generators
generators = Generators()
# generate 8 samples with model 8 (00008_C-DCGAN_MMG_MASSES).
# Also, auto-install required model dependencies.
generators.generate(model_id=8, num_samples=8, install_dependencies=True)The synthetic images in the top row show malignant masses (breast cancer) while the images in the bottom row show benign masses. Given such images with class information, image classification models can be (pre-)trained.
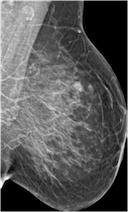
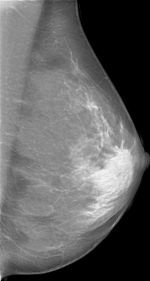
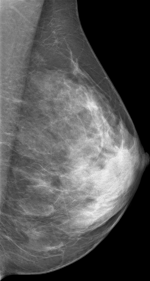
Create mammograms translated from Low-to-High Breast Density using CYCLEGAN model
from medigan import Generators
generators = Generators()
# model 3 is "00003_CYCLEGAN_MMG_DENSITY_FULL"
generators.generate(model_id=3, num_samples=1)Search for a model inside medigan using keywords
# import medigan and initialize Generators
from medigan import Generators
generators = Generators()
# list all models
print(generators.list_models())
# search for models that have specific keywords in their config
keywords = ['DCGAN', 'Mammography', 'BCDR']
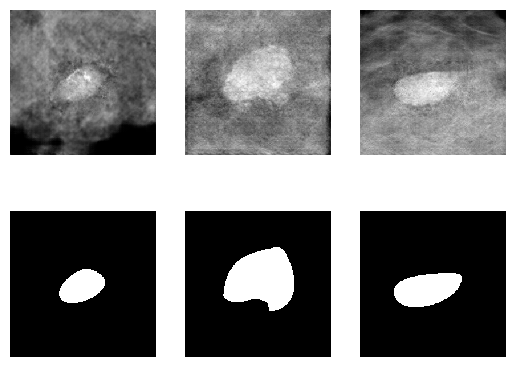
results = generators.find_matching_models_by_values(keywords)We can directly receive a torch.utils.data.DataLoader object for any of medigan's generative models.
from medigan import Generators
generators = Generators()
# model 4 is "00004_PIX2PIX_MMG_MASSES_W_MASKS"
dataloader = generators.get_as_torch_dataloader(model_id=4, num_samples=3)Visualize the contents of the dataloader.
from matplotlib import pyplot as plt
import numpy as np
plt.figure()
# subplot with 2 rows and len(dataloader) columns
f, img_array = plt.subplots(2, len(dataloader))
for batch_idx, data_dict in enumerate(dataloader):
sample = np.squeeze(data_dict.get("sample"))
mask = np.squeeze(data_dict.get("mask"))
img_array[0][batch_idx].imshow(sample, interpolation='nearest', cmap='gray')
img_array[1][batch_idx].imshow(mask, interpolation='nearest', cmap='gray')
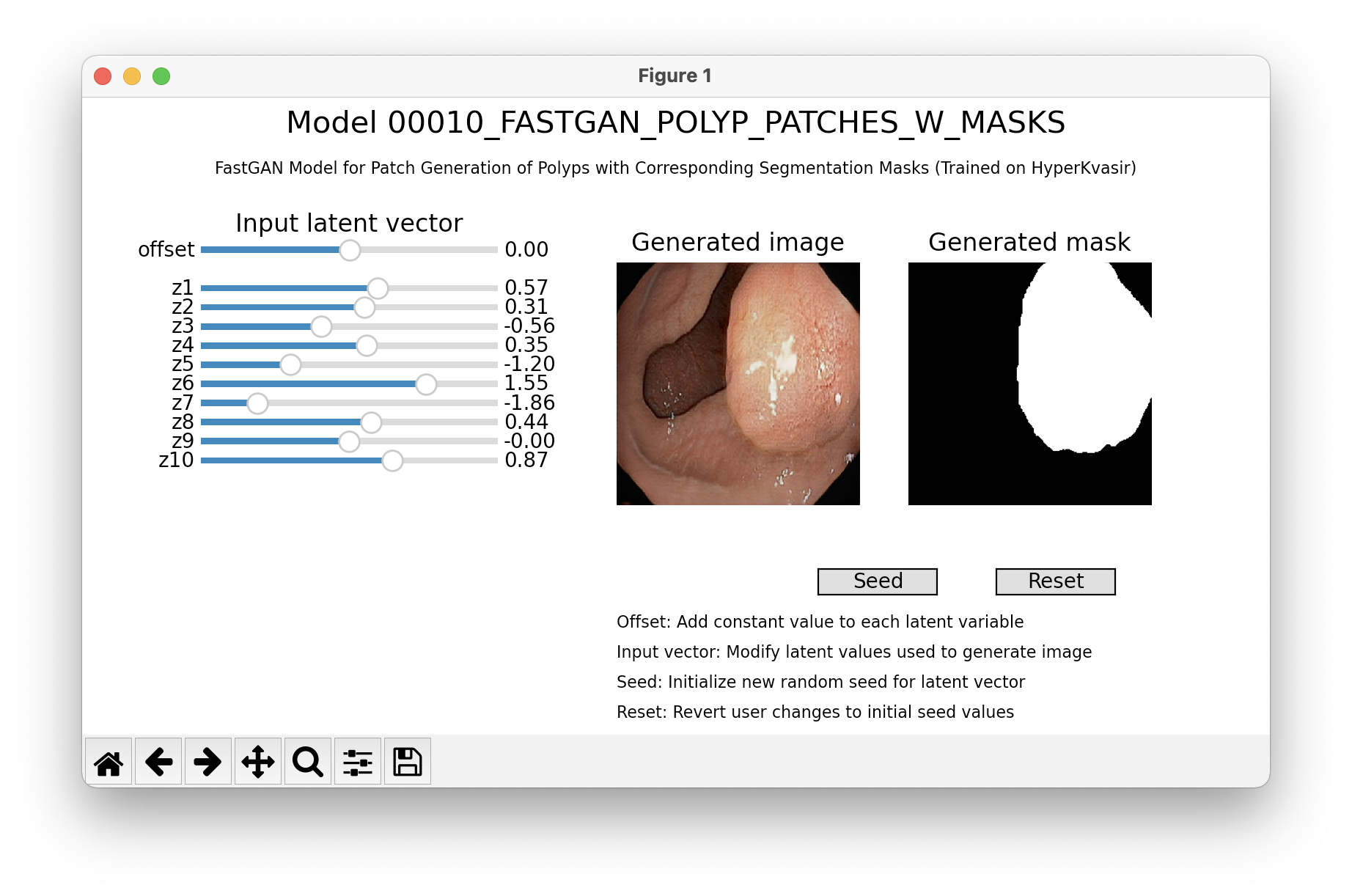
plt.show()With our interface, it is possible to generate sample by manually setting the conditional inputs or latent vector values. The sample is updated in realtime, so it's possible to observe how the images changes when the parameters are modified. The visualization is available only for models with accessible input latent vector. Depending on a model, a conditional input may be also available or synthetic segmentation mask.
from medigan import Generators
generators = Generators()
# model 10 is "00010_FASTGAN_POLYP_PATCHES_W_MASKS"
generators.visualize(10)Create an init.py file in your model's root folder.
Next, run the following code to contribute your model to medigan.
-
Your model will be stored on Zenodo.
-
Also, a Github issue will be created to add your model's metadata to medigan's global.json.
-
To do so, please provide a github access token (get one here) and a zenodo access token (get one here), as shown below. After creation, the zenodo access token may take a few minutes before being recognized in zenodo API calls.
from medigan import Generators
generators = Generators()
# Contribute your model
generators.contribute(
model_id = "00100_YOUR_MODEL", # assign an ID
init_py_path ="path/ending/with/__init__.py",
model_weights_name = "10000",
model_weights_extension = ".pt",
generate_method_name = "generate", # in __init__.py
dependencies = ["numpy", "torch"],
creator_name = "YOUR_NAME",
creator_affiliation = "YOUR_AFFILIATION",
zenodo_access_token = 'ZENODO_ACCESS_TOKEN',
github_access_token = 'GITHUB_ACCESS_TOKEN',Thank you for your contribution!
You will soon receive a reply in the Github issue that you created for your model by running generators.contribute().
We welcome contributions to medigan. Please send us an email or read the contributing guidelines regarding contributing to the medigan project.
If you use a medigan model in your work, please cite its respective publication (see references).
Please also consider citing the medigan paper:
BibTeX entry:
@article{osuala2023medigan,
title={medigan: a Python library of pretrained generative models for medical image synthesis},
author={Osuala, Richard and Skorupko, Grzegorz and Lazrak, Noussair and Garrucho, Lidia and Garc{\'\i}a, Eloy and Joshi, Smriti and Jouide, Socayna and Rutherford, Michael and Prior, Fred and Kushibar, Kaisar and others},
journal={Journal of Medical Imaging},
volume={10},
number={6},
pages={061403},
year={2023},
publisher={SPIE}
}