Clustering motif models to remove redundancy
- Python 2.7
- numpy, scipy
- genome-tools (http://www.github.com/jvierstra/genome-tools)
- Tomtom (http://meme-suite.org/doc/download.html)
- Jolma et al., Cell 2013 (Supplemental Table 2)
- JASPAR 2018
- HOCOMOCO version 11 (757 motif models; both human and mouse)
We have pre-computed genome-wide scans for both human and mouse genomes.
- Human (GRCh38/h38)
- Full motif matches (2179 motif models)
- Collapsed motifs by similarity (286 motif clusters)
- Mouse (mm10)
- Full motif matches (2179 motif models)
- Collapsed motifs by similarity (286 motif clusters)
Here we TOMTOM to determine the similarity between all motif models (all pairwise) with the following code:
meme2meme databases/*/*.meme > tomtom/all.dbs.meme
tomtom \
-bfile /net/seq/data/projects/motifs/hg19.K36.mappable_only.5-order.markov \
-dist kullback \
-motif-pseudo 0.1 \
-text \
-min-overlap 1 \
tomtom/all.dbs.meme tomtom/all.dbs.meme \
> tomtom/tomtom.all.txt
I have a provided a script that will load this operation up on a SLURM parallel compute cluster (see e)
python2 hierarchical.py tomtom/tomtom.all.txt tomtom
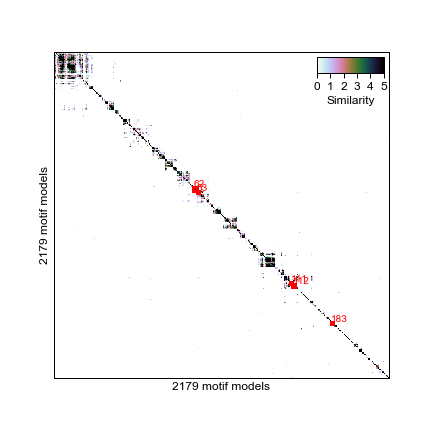
This script performs hierarchical clustering (distance: correlation, complete linkage) and provides an output of cluster assignments at a range of tree heights (0.5-1). Below is a heatmap representation of motifs clustered by simililarity and clusters identified cutting the dendrogram at height 0.7.
mkdir -p tomtom/height.0.70/viz
python2 process_cluster.py \
tomtom/tomtom.all.txt \
tomtom/clusters.0.70.txt \
62 \
tomtom/height.0.70
This command generates two files (per motif cluster).
python2 viz_cluster.py \
tomtom/height.0.70/cluster-info.62.txt \
tomtom/height.0.70/cluster-motifs.62.txt \
tomtom/height.0.70/viz/cluster.62
This wiil create a PDF and PNG with visualizing motif cluster #62 corresponding to the basic helix-loop-helix DBD containing OLIG/NEUROG. Dashed lines demarcate the boundaries of the "archetypal" motif position. The motif matches for the constituent models have will have their coordinates adjusted to match.
| C62:OLIG (bHLH) | C179:RUNX (RUNX domain) |
|---|---|
 |
 |
fetchChromSizes hg38 > /tmp/chrom.sizes
awk -v OFS="\t" '{ print $1, 0, $2; }' /tmp/chrom.sizes | sort-bed - > /tmp/chrom.sizes.bed
zcat moods.combined.all.bed.gz | bedops -e 100% - /tmp/chrom.sizes.bed \
| python2 /home/jvierstra/proj/code/motif-clustering/relabel.py \
/home/jvierstra/proj/code/motif-clustering/tomtom/height.0.70/cluster-info.with.dbd.and.color.csv \
| head -n10000 > /tmp/moods
To create a bigBed file from a bed9+2, we need to include an AutoSql file (bed_format.as)
table hg38_motifs_collapsed
"Collapsed motifs matches in hg38 (see: http://www.github.com/jvierstra/motif-clustering)"
(
string chrom; "Reference sequence chromosome or scaffold"
uint chromStart; "Start position of feature on chromosome"
uint chromEnd; "End position of feature on chromosome"
string name; "Name of gene"
uint score; "Score"
char[1] strand; "+ or - for strand"
uint thickStart; "Coding region start"
uint thickEnd; "Coding region end"
uint reserved; "itemRgb"
string motif_match_id; "Motif identifier"
float motif_match_score; "Motif match score (MOODS score)"
string DBD; "DNA binding domain"
uint n; "Number of motif matches from cluster"
)
bedToBigBed -as=bed_format.as -type=bed9+4 -tab moods.combined.all.bed chrom.sizes moods.combined.all.bb
awk -v OFS="\t" '{ print $1, $2, $3, $4, $11, $6, $10, $13}' moods.combined.all.bed | bgzip -c > moods.combined.all.bed.gz
tabix -p bed moods.combined.all.bed.gz
awk -v OFS="\t" '{ print $1, $2, $3, $4, 0, $6, $2, $3, "0,0,0", $5, $7 }' moods.combined.all.bed > /tmp/moods &