Python lib for common ontology operations over a variety of backends.
OAK provides a collection of interfaces for various ontology operations, including:
- look up basic features of an ontology element, such as its label, definition, relationships, or aliases
- search an ontology for a term
- validate an ontology
- modify or delete terms
- generate and visualize subgraphs
- identify lexical matches and export as SSSOM mapping tables
- perform more advanced operations, such as graph traversal, OWL axiom processing, or text annotation
These interfaces are separated from any particular backend, for which there a number of different adapters. This means the same Python API and command line can be used regardless of whether the ontology:
- is served by a remote API such as OLS or BioPortal
- is present locally on the filesystem in owl, obo, obojson, or sqlite formats
- is to be downloaded from a remote repository such as the OBO library
- is queried from a remote database, including SPARQL endpoints (Ontobee/Ubergraph), A SQL database, a Solr/ES endpoint
- incatools.github.io/ontology-access-kit
- Presentations:
- Using the OAK command line OBO Academy 2023
- Introduction to OAK OAK workshop 2022
See the contribution guidelines at CONTRIBUTING.md. All contributors are expected to uphold our Code of Conduct.
from oaklib import get_adapter
# connect to the CL sqlite database adapter
# (will first download if not already downloaded)
adapter = get_adapter("sqlite:obo:cl")
NEURON = "CL:0000540"
print('## Basic info')
print(f'ID: {NEURON}')
print(f'Label: {adapter.label(NEURON)}')
for alias in adapter.entity_aliases(NEURON):
print(f'Alias: {alias}')
print('## Relationships (direct)')
for relationship in adapter.relationships([NEURON]):
print(f' * {relationship.predicate} -> {relationship.object} "{adapter.label(relationship.object)}"')
print('## Ancestors (over IS_A and PART_OF)')
from oaklib.datamodels.vocabulary import IS_A, PART_OF
from oaklib.interfaces import OboGraphInterface
if not isinstance(adapter, OboGraphInterface):
raise ValueError('This adapter does not support graph operations')
for ancestor in adapter.ancestors(NEURON, predicates=[IS_A, PART_OF]):
print(f' * ANCESTOR: "{adapter.label(ancestor)}"')For more examples, see
See:
Use the pronto backend to fetch and parse an ontology from the OBO library, then use the search command
runoak -i obolibrary:pato.obo search osmol Returns:
PATO:0001655 ! osmolarity
PATO:0001656 ! decreased osmolarity
PATO:0001657 ! increased osmolarity
PATO:0002027 ! osmolality
PATO:0002028 ! decreased osmolality
PATO:0002029 ! increased osmolality
PATO:0045034 ! normal osmolality
PATO:0045035 ! normal osmolarity
Perform validation on PR using sqlite/rdftab instance:
runoak -i sqlite:../semantic-sql/db/pr.db validateList all terms obolibrary has for mondo
runoak -i obolibrary:mondo.obo terms Make a lexical index of all terms in Mondo:
runoak -i obolibrary:mondo.obo lexmatch -L mondo.index.yamlSearching over OBO using ontobee:
runoak -i ontobee: search tentacleyields:
http://purl.obolibrary.org/obo/CEPH_0000256 ! tentacle
http://purl.obolibrary.org/obo/CEPH_0000257 ! tentacle absence
http://purl.obolibrary.org/obo/CEPH_0000258 ! tentacle pad
...
Searching over a broader set of ontologies in bioportal (requires API KEY) (https://www.bioontology.org/wiki/BioPortal_Help#Getting_an_API_key)
runoak set-apikey bioportal YOUR-KEY-HERE
runoak -i bioportal: search tentacleyields:
BTO:0001357 ! tentacle
http://purl.jp/bio/4/id/200906071014668510 ! tentacle
CEPH:0000256 ! tentacle
http://www.projecthalo.com/aura#Tentacle ! Tentacle
CEPH:0000256 ! tentacle
...
Alternatively, you can add "BIOPORTAL_API_KEY" to your environment variables.
Searching over more limited set of ontologies in Ubergraph:
runoak -v -i ubergraph: search tentacleyields
UBERON:0013206 ! nasal tentacle
runoak -i bioportal: annotate neuron from CA4 region of hippocampus of mouseyields:
object_id: CL:0000540
object_label: neuron
object_source: https://data.bioontology.org/ontologies/NIFDYS
match_type: PREF
subject_start: 1
subject_end: 6
subject_label: NEURON
object_id: http://www.co-ode.org/ontologies/galen#Neuron
object_label: Neuron
object_source: https://data.bioontology.org/ontologies/GALEN
match_type: PREF
subject_start: 1
subject_end: 6
subject_label: NEURON
...Create a SSSOM mapping file for a set of ontologies:
robot merge -I http://purl.obolibrary.org/obo/hp.owl -I http://purl.obolibrary.org/obo/mp.owl convert --check false -o hp-mp.obo
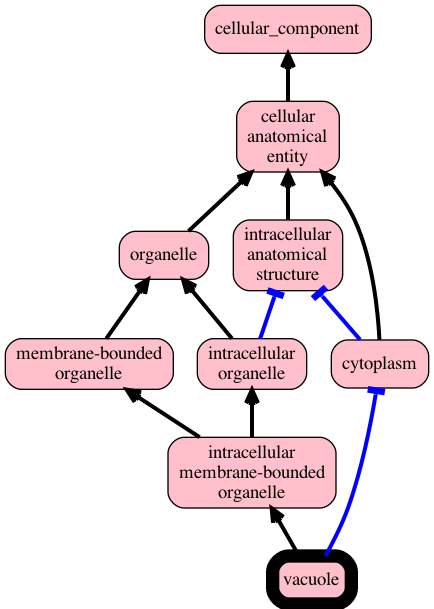
runoak lexmatch -i hp-mp.obo -o hp-mp.sssom.tsvUse the sqlite backend to visualize graph up from 'vacuole' using test ontology sqlite:
runoak -i sqlite:tests/input/go-nucleus.db viz GO:0005773Same using ubergraph, restricting to is-a and part-of
runoak -i ubergraph: viz GO:0005773 -p i,BFO:0000050Same using pronto, fetching ontology from obolibrary
runoak -i obolibrary:go.obo viz GO:0005773OAK uses pystow for caching. By default,
this goes inside ~/.data/, but can be configured following
these instructions.