TEDAR is a pharmacovigilance signal detection method based on variable-length temporal splitting.
The main goal is to detect, for each specific drug-adr pair, a set of intervals having different lengths that are representative of the pair under consideration. A set of overlapping intervals are extracted for each drug-adr pair by applying a temporal data-mining approach. The notion of homogeneous interval is introduced. The covariance coefficient is engaged for detecting cutting points between the intervals in order to extract only homogeneous intervals. Then, a graph theory-based algorithm is applied for retrieving a final set of non-overlapping intervals. Finally, TEDAR uses the PRR statistics for evaluating the significance of the retrieved intervals.
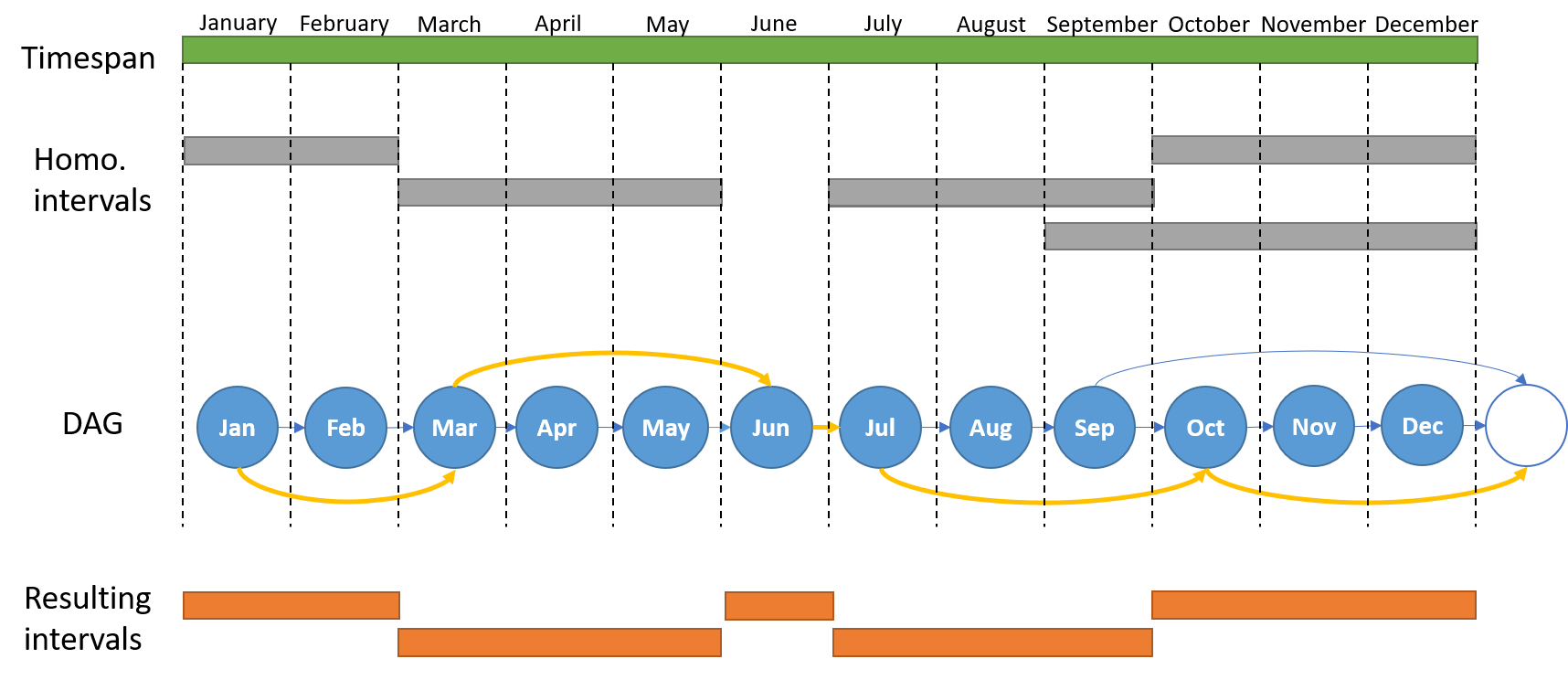
The above image represents the generation of a DAG of intervals for extracting non-overlapping homogeneous intervals within the timespan of a specific drug-adr pair. Starting homogeneous intervals are displayed as grey rectangles. The initial structure of the DAG is the set of ordered time points (in this case months), represented as nodes (blue nodes) and linked by single edges (blue edges). Initial intervals are embedded in the DAG by adding extra edges from the starting time point to the DAG node consecutive to the one representing the end of the interval (blu edges). For this reason, an extra node is queued to the DAG (white node) in order to represent intervals in which the endpoint is the end of the timespan. Final intervals (orange rectangles) are extracted from one of the possible shortest paths (yellow path) from the start to the end of the DAG.
TEDAR is released in a Docker container, that allows to isolate applications from their environment, with the effect of increasing replicability. All dependencies are automatically installed when the container is created (see TEDAR/DockerContainer/TEDAR/Dockerfile).
The TEDAR software is developed using Ruby scripts in a set of Jupyter Notebooks. Reports are stored and manipulated by using Redis as database management system.
R scripts are developed for applying the signal detection thresholds and the validation phase of drug-adrs detected.
We use as a case study the surveillance database, named RNF (Rete Nazionale Farmacovigilanza), released by the Italian authority AIFA (Agenzia Italiana del Farmaco). The RNF database contains reports of ADRs issued by all the Italian regions.
ADRs are encoded according to the MedDRA (Medical Dictionary for Regulatory Activities) terminology, which consists of a large set of terms structured into five hierarchical levels. System Organ Classes (SOC) are the level terminology used in this system to encode ADRs. SOC is the highest level of ADR terminology and terms here are distinguished by anatomical or physiological system, etiology or purpose.
Drug is defined as a pharmaceutical product (combinations of active ingredients) according to the requirements of the ICH M5 standard adopted in RNF. We make no distinction between pharmaceutical products with the same combinations of active ingredients.
Data extraction from RNF was carried out through the Vigisegn data warehouse.
We used the ADReCS and PROTECT datasets containing verified drug-adr relations for assessing the performances of TEDAR. The reference dataset used is obtained by merging these two datasets. Furthermore we selected only the drug-ard pairs for which a minimum number of reports equal to 5 is reported in RNF. The excluded pairs did not have enough support in the RNF dataset to be detected as signals.
Input data and reference dataset provided in this repository are an anonimyzed version of RNF, thus contain drugs encoded as: drug1, drug2, ... drug3042 .
Reference dataset reference_dataset.txt is contained in the DockerContainer/TEDAR directory.
The complete set of reports must be provided as a text file. The file contains one report per line represented as a date of insertion, a drug and an adr. Fields in a record are separated by tabs.
A valid file is given by the following example:
vt drug soc
2017-01-02 drug169 Gastrointestinal disorders
2017-01-02 drug169 Vascular disorders
2017-01-02 drug169 Musculoskeletal and connective tissue disorders
2017-01-02 drug169 Blood and lymphatic system disorders
2017-01-11 drug170 Blood and lymphatic system disorders
2017-01-18 drug171 General disorders and administration site conditions
2017-01-20 drug172 Investigations
2017-01-20 drug172 Blood and lymphatic system disorders
2017-01-20 drug172 Hepatobiliary disorders
2017-01-23 drug32 General disorders and administration site conditions
2017-01-23 drug130 Skin and subcutaneous tissue disorders
2017-01-23 drug130 Gastrointestinal disorders
2017-01-23 drug130 Vascular disorders
2017-01-23 drug130 Gastrointestinal disorders
2017-01-23 drug130 Nervous system disorders
2017-01-23 drug158 General disorders and administration site conditions
2017-02-06 drug173 Skin and subcutaneous tissue disorders
The input file must be specified in Init.ipynb (INPUTDATA constant). It is necessary to modify START_MONTH and END_MONTH in TEDAR.ipynb and Compute_disprortionality.pynb source code according to the timespan to be analyzed, i.e. timespan from 2008-1-1 to 2017-12-1 required START_MONTH=[2008,1] and END_MONTH=[2017,12] ([year, month]).
In the DockerContainer/TEDAR/sciruby/ folder there are two encoded versions of the input data (the requested time to import the first input file to Redis is about 1 hour, for the second one is about 7 hours - times refers to a laptop with XXXXX)
input_data_1y.txt: encoded reports in collected in RNF in 2017;input_data_10.rar: encoded reports in collected in RNF in [2008,2017] (extract the .rar file);
The TEDAR version provided in this repository uses input_data_1y.txt as default input. To use input_data_10y.txt see comments in Init.ipynb (INPUTDATA constant), TEDAR.ipynb (START_MONTH and END_MONTH constant), and Compute_disprortionality.ipynb (START_MONTH and END_MONTH constant).
Docker is required.
The user has also to ensure that Docker is currently installed and there are no too strict limits on the number of CPUs and amount of memory that Docker can use (https://docs.docker.com/config/containers/resource_constraints/ for further details).
Download and extract the repository, then move to DockerContainer/TEDAR/ and run from terminal:
docker-compose up
To execute the code inside the Jupyter Notebook open http://localhost:8888/ via broswer.
Source code is provided in DockerContainer/TEDAR/sciruby/.
The 3 ipynb files can be easly run in Jupyter Notebook via graphical interface. It is recommended to run the files in this order:
Init.ipynbTEDAR.ipynbCompute_disproportionality.ipynb
File needed to upload input data in Redis database.
Input data must be provided as specified in Input Data. Set INPUTDATA constant to specify the path.
This file is the core file of TEDAR methodology.
Given the input data already uploaded in Redis database, homogenous intervals are obtained and written to the file DockerContainer/TEDAR/sciruby/results/TEDAR/split/split_TEDAR.txt.
split_TEDAR.txt is a tab separated text file that contains the homogenous intervals for each drug-adr pair in a line.
Here an example listing 3 drug-adr pairs:
drug166 Product issues 0,1,13
drug429 Skin and subcutaneous tissue disorders 0,1,3,5,8,9,13
drug202 Blood and lymphatic system disorders 0,7,10,13
Set START_MONTH and END_MONTH to specify the timespan to be analyzed, i.e. timespan from 2008-1-1 to 2017-12-1 required START_MONTH=[2008,1] and END_MONTH=[2017,12] ([year, month]).
This file computes PRR and metrics applied according the thresholds (Confidence Interval and Chi-squared statistics).
Set START_MONTH and END_MONTH to specify the timespan to be analyzed, i.e. timespan from 2008-1-1 to 2017-12-1 required START_MONTH=[2008,1] and END_MONTH=[2017,12] ([year, month]).
There are 4 methodologies that can be runned varying the time unit: TEDAR (variable length intervals), PRR monthly (1 month length intervals), PRR quarterly (3 months length intervals), PRR yearly (annual length intervals).
TEDAR analysis requires the generation of split_TEDAR.txt as described in TEDAR.ipynb.
For each methodology, a file in results directory reports the obtained metrics (DockerContainer/TEDAR/sciruby/results/TEDAR/result_TEDAR.txt, DockerContainer/TEDAR/sciruby/results/TEDAR/result_prr_monthly.txt, DockerContainer/TEDAR/sciruby/results/TEDAR/result_prr_quarterly.txt, DockerContainer/TEDAR/sciruby/results/TEDAR/result_prr_yearly.txt).
Output file is a tab separated text file containing a line for each interval of the analysed drg-adr pairs:
Drug Adr Start_month End_month Prr LowerBoundConfidenceInterval UpperBoundConfidenceInterval Chi-squared NumberOfReportInIntervals
An example of output file is showed in the following lines listing results for pairs "drug166-Product issues" and "drug289-Gastrointestinal disorders" using TEDAR (variable lenght intervals) in timespan [2017-1-1,2017-12-31]:
drug166 Product issues [2017, 1] [2017, 3] 13.567421790722761 5.702329665607122 32.28065454679049 51.50491267314168 5
drug166 Product issues [2017, 4] [2017, 6] 0.0 0.0 NaN 0.19328512034182097 0
drug166 Product issues [2017, 7] [2017, 9] 0.0 0.0 NaN 0.09696418479286524 0
drug166 Product issues [2017, 10] [2017, 12] 0.0 0.0 NaN 0.4265674326620676 0
drug289 Gastrointestinal disorders [2017, 1] [2017, 3] 0.13697869244542418 0.01964111254018487 0.9553003754583395 5.470147794547452 1
drug289 Gastrointestinal disorders [2017, 4] [2017, 6] 0.8410845847520452 0.33146707846440043 2.1342188249428142 0.30023831513894006 4
drug289 Gastrointestinal disorders [2017, 7] [2017, 9] 1.2641056422569028 0.6881028636443149 2.322273542537871 0.033651415267548335 9
drug289 Gastrointestinal disorders [2017, 10] [2017, 12] 0.5401822700911351 0.20967439923221448 1.3916667270268261 1.8536783809609447 4
Submitted.