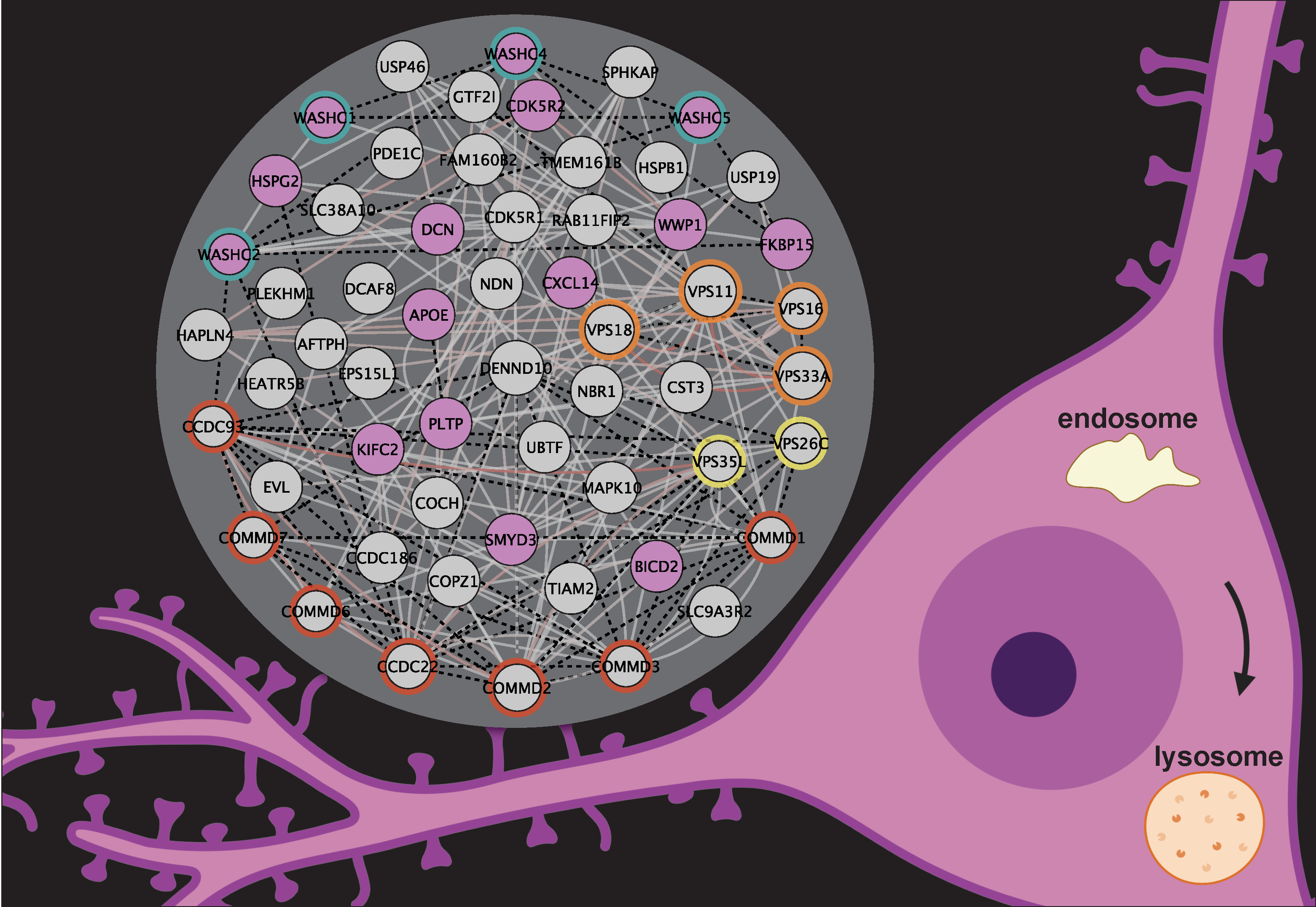
This repository contains the raw TMT and BioID proteomics data associated with the analyses performed by Courtland et al., 2021 [1].
The analysis utilizes functions from
soderling-lab/tidyProt to perform
protein- and module- level comparisons (see example below). Key data and
results are saved as R objects in the data/ directory. These can be accessed in R using the
data() function. For example, load the Swip spatial proteome network partition with
data(swip_partition, package="SwipProteomics").
The statistical results for protein- and module-level analyses can be found in
the tables/ directory.
Maps for plasmids used by this study are in the sequences/ directory.
A Cytoscape graph of the Swip spatial proteome partitioned into 49 modules using network enhancement [2] and the leiden algorithm [3] is available in inst/extdata.
# download the repository as an R package
devtools::install_github("soderling-lab/SwipProteomics")
# install tidyProt for statistical functions
devtools::install_github("soderling-lab/tidyProt")
library(dplyr)
library(tidyProt)
library(SwipProteomics)
# load the normalized TMT data
data(swip_tmt)
# washc4's uniprot ID
data(swip)
# fit a model
fx <- log2(Intensity) ~ 0 + Condition + (1|Mixture)
fm <- lmerTest::lmer(fx,data=swip_tmt %>% subset(Protein==swip))
# create a contrast
LT <- getContrast(fm,"Mutant","Control")
# assess contrast
res <- lmerTestContrast(fm, LT) %>% mutate(Contrast='Mutant-Control') %>% unique()
knitr::kable(res)
| Contrast | log2FC | percentControl | SE | Tstatistic | Pvalue | DF | S2 | isSingular |
|---|---|---|---|---|---|---|---|---|
| Mutant-Control | -1.401866 | 0.3784393 | 0.0264791 | -52.94235 | 0 | 28 | 0.007362 | TRUE |
lmerTestContrast returns a data.frame with statistics from the model-based
contrast. The column isSingular=TRUE in this case indicates that the variance
attributes to Mixture is negligible.
## fit WASH Complex
library(dplyr)
library(SwipProteomics)
data(washc_prots)
# module-level model includes ranef Protein
fx1 <- log2(rel_Intensity) ~ 0 + Condition + (1|Protein)
# fit the model
fm1 <- lmerTest::lmer(fx1, data = swip_tmt %>% subset(Protein %in% washc_prots))
# assess overall 'Mutant-Control' comparison
res1 <- lmerTestContrast(fm1, LT) %>% mutate(Contrast='Mutant-Control') %>% unique()
knitr::kable(res)
| Contrast | log2FC | percentControl | SE | Tstatistic | Pvalue | DF | S2 | isSingular |
|---|---|---|---|---|---|---|---|---|
| Mutant-Control | -1.379633 | 0.3843165 | 0.0392109 | -35.18497 | 0 | 151 | 0.0645747 | FALSE |
@article{Courtland2021,
title = {Genetic Disruption of WASHC4 Drives Endo-lysosomal Dysfunction and Cognitive-Movement Impairments in Mice and Humans},
author = {Jamie L. Courtland and Tyler W. A. Bradshaw and Greg Waitt and Erik J. Soderblom and Tricia Ho and Anna Rajab and
Ricardo Vancini and Il Hwan Kim and Scott H. Soderling},
year = 2021,
month = {March},
journal = {eLife},
doi = {https://doi.org/10.7554/eLife.61590},
url = {none}
}
[1] Genetic Disruption of WASHC4 Drives Endo-lysosomal Dysfunction and
Cognitive-Movement Impairments in Mice and Humans.
Courtland J.L., Bradshaw T.W.A., Waitt G., Soderblom E., Ho T., Rajab A.,
Vancini R., Kim I.H., Soderling S.H. (2021). eLife; 10:e61590
doi: 10.7554/eLife.61590
[2] From Louvain to Leiden: guaranteeing well-connected communities.
Traag, V.A., Waltman. L., Van Eck, N.-J. (2018). Scientific reports, 9(1), 5233.
10.1038/s41598-019-41695-z
[3] Network Enhancement as a general method to denoise weighted biological networks.
Wang B., Pourshafeie A., Zitnik M., Zhu J., Bustamante C.D., Batzoglou S., Leskovec J. (2018).
Nature Communications, 9, 3108. 10.1038/s41467-018-05469-x