FireCore is an integrated simulation environment dedicated to on-surface chemistry and scanning-probe microscopy (SPM). The code aims to streamline high-throughput screening of molecular configurations and processes on the surface of inorganic crystals, including:
- exploration of non-covalent self-assembling processes
- exploration of chemical reaction pathways between organic molecules on surface
- computational prototyping of molecular designs for pre-programmed self-assembling
- simulation of high-resolution AFM / STM experiments of organic molecules
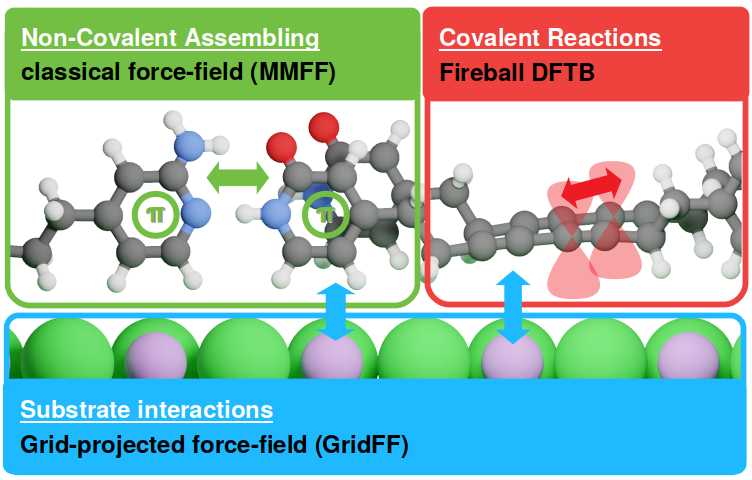
To achieve a favorable compromise between speed and accuracy the code use combination of both ab-initio quantum methods and classical forcefield (QM/MM approach) and extensive GPU acceleration.
FireCore consists of high-performance simulation modules written in Fortran, C/C++ & OpenCL integrated using a common Python3 interface. The modules are based on a modified version of other simulation packages.
Striped down version of Fireball self-consistent local-orbital ab-initio tight-binding molecular dynamics code (https://github.com/fireball-QMD/progs) is used for the following tasks:
To optimize the electronic structure of molecules and find molecular orbitals and density To optimize the geometric structure of molecules (quantum part of QM/MM)
- To project Molecular Orbitals and Electron density to a real space grid
Fireball allows the fast solution of self-consistent electronic structure problems thanks to a small optimized numerical base and pre-calculated table of integrals.
FireCore is currently integrated with homebrew classical-forcefields codes implemented in SimpleSimulationEngine (https://github.com/ProkopHapala/SimpleSimulationEngine). The main purpose is to reduce the computational cost required for the relaxation of organic molecules and the description of the interaction between organic molecules and substrate. These codes comprise:
- MMFF - a simple homebrew classical molecular mechanics code with fixed boding topology (springs on bond length, angles and dihedrals)
- Code includes utilities to automatically generate bonding topology from atomic coordinates and assign atomic charges by the charge-equilibration algorithm (depending on electronegativity and local polarization) RFF - simple homebrew reactive force-field to roughly simulate chemical interactins between organic species (H,C,O,N) including formation and breaking of new bonds and changes of bond-order gridFF - grid-based classical forcefield to describe the interaction of organic molecules with the rigid substrate or other rigid bodies (e.g. tip of AFM?). The code projects non-covalent forces from rigid objects (e.g. electrostatics, van der Waals attraction and Pauli repulsion) onto a real-space grid. Then during molecular-dynamics simulation, we read from the grid forces acting on each atom of the flexible (non-rigid) molecules deposited on the rigid substrate.
In future we plan integration with state-of-the-art classical forcefield packages such as LAMMPS.
GPU accelerated Probe Particle Model (https://github.com/ProkopHapala/ProbeParticleModel) is used for the simulation of high-resolution AFM images using flexible tip (e.g. CO molecule or Xe atom). It was shown that variation of local electrostatic field and electron density (e.g. bond-order and free electron pairs) plays an important role in the formation of HR-AFM contrast and its distortions. To obtain a good estimate of electron density and electrostatic potential we use a self-consistent electronic structure calculated using fireball as an input. With a combination of classical forcefield relaxation of molecular geometry, fireball solution of self-consistent electronic structure and GPU accelerated AFM relaxation using FireCore is able to conduct a series of volumetric AFM simulations (starting from molecular topology) in less than 1 second. This allows the rapid screening of candidate molecular structures to match experimental data, or to rapidly generate large databases for training machine-learning models for AFM imaging.
We use GPU acceleration using OpenCL kernels for most performance-intensive steps of the simulation, including:
- Simulation of AFM scan where probe-particle (CO apex) relax in Volumgetric potential (gridFF) comprising of electrostatics, van der Waals attraction and Pauli repulsion from the rigid sample.
- Evaluation of gridFF by the projection of contributions from each atom of the sample - comprising van der Waals attraction and Pauli.
- Evaluation of electrostatic potential and forcefield on the grid from electron density (i.e. solution of Poisson equation) using Fast-Fourier-Transform. We use library clFFT for the purpose. Projection of wave-functions or electron density from FireBall onto real space grid using FireBall numerical basis.
To install all library dependencies. On Ubuntu 22.04 this can be done by running:
sudo apt-get install cmake g++ gfortran intel-mkl libmkl_intel_lp64 libmkl_intel_thread libsdl2-dev libsdl2-image-dev nvidia-opencl-dev libclfft-dev python3-numpy python3-matplotlib
however individual parts can be installed independently. For example, it is possible to use DFT-Fireball without OpenCL or OpenGL.
- install Fortran compiler:
gfortran - install intel mkl libraries and other dependencies:
intel-mkl libmkl_intel_lp64 libmkl_intel_core libmkl_intel_thread - create directory
FireCore/Build - run
FireCore/make.shwhich will compile source codes fromfortrandirectory intoBuilddirectory - run test:
- navigate to
FireCore/tests - Make sure that
Fdata_HC_minimalis present, otherwise download it here: https://fireball-qmd.github.io/ - got to
tests/t01_H2and run./run.sh
- navigate to
- install C/C++ compiler:
g++ cmake - install libraries:
- for GUI with SDL2+OpenGL install:
libsdl2-dev libsdl2-image-dev - for GPU accelerated calculation with OpenCL install:
nvidia-opencl-dev libclfft-devcreate a build directory incpp/Buildand navigate into it
- for GUI with SDL2+OpenGL install:
- Configure cmake project: from inside
cpp/Buildruncmake .. -DWITH_SDL=ON -DWITH_OPENCL=ON- options
-DWITH_SDL=ONand-DWITH_OPENCL=ONcan be eventually switched to reduce compilation time and/or limit library dependencies
- options
- compile by running
makeinsidecpp/Builddirectory - Run tests:
- Forcefield with visual GUI go to
/home/prokophapala/git/FireCore/cpp/sketches_SDL/Molecularand run e.g../test_MMFFsp3ortest_RARFFarrortest_SoftMolecularDynamics - to run Visual GUI for QMMM dynamics (assuming both C++ and Fortran modules are compiled) got to
FireCore/tests/tQMMM_diacetyleneand run./run.sh
- Forcefield with visual GUI go to
- Try classical forcefield library
MMFFsp3go tpFireCore/tests/tMMFFand run./run.sh - Try Fireball-DFT in
FireCore/tests/FitFFby running./run.sh