Harmonization tools for multi-site neuroimaging analysis. Part of the work reported in our first paper with data from the ISTAGING consoritum [1].
This package extends the functionality of the package developed by Nick Cullen [2],
neuroCombat, which is hosted on GitHub: https://github.com/ncullen93/neuroCombat
The previously-released package, neuroCombat, allows the user to perform a
harmonization procedure using the ComBat [3] algorithm for correcting
multi-site data.
This package, neuroHarmonize, has similar functionality, but also allows the
user to perform additional procedures:
- Train a harmonization model on a subset of data, then apply the model to the new set. For example, in longitudinal analyses, one may wish to train a harmonization model on baseline cases and apply the model to follow-up cases, to avoid double-counting subjects.
- Specify covariates with nonlinear effects. Age tends to exhibit nonlinear
relationships with brain volumes. Nonlinear effects are implemented using
Generalized Additive Models (GAMs) via the
statsmodelspackage. - Apply a pre-trained harmonization model to NIFTI images. When performing
image-level harmonization, loading the entire set of images may exceed
memory capacity. In such cases, it is still possible to harmonize images by
sequentially adjusting images one-by-one. This functionality is made
available via the
nibabelpackage. - Train a harmonization model without the empirical Bayes (EB) step of ComBat.
Latest version: 0.3.1 (April 2020)
Requirements:
git >= 2.17.2python >= 3.7.6
To make installation easier, neuroCombat is not a formal dependency for this package, but the source code is included to call neuroCombat functions.
Option 1: Install from PyPI (not available yet)
Instructions will be written once package is released on PyPI. The package cannot be released on PyPI until a developer version of statsmodels is released.
Option 2: Install from GitHub
Install the developer version of
statsmodels. This package depends onstatsmodels v0.12.0.dev0. Until the dev version is released, the current workaround is to run the following in the command line:>>> pip install git+https://github.com/statsmodels/statsmodels
Install latest version of
neuroHarmonize. Run the following in the command line:>>> pip install git+https://github.com/rpomponio/neuroHarmonize
You must provide a data matrix which is a numpy.array containing the
features to be harmonized. For example, an array of brain volumes:
array([[3138.0, 3164.2, ..., 206.4],
[1708.4, 2351.2, ..., 364.0],
...,
[1119.6, 1071.6, ..., 326.6]])
The dimensionality of this matrix must be: N_samples x N_features
You must also provide a covariate matrix which is a pandas.DataFrame
containing all covariates to control for during harmonization. All covariates
must be encoded numerically (you must handle categorical covariates in a
pre-processing step, see pandas.get_dummies). The DataFrame must
also contain a single column called "SITE" with labels that identify sites
(the labels in "SITE" need not be numeric).
SITE AGE SEX_M 0 SITE_A 76.5 1 1 SITE_B 80.1 1 2 SITE_A 82.9 0 ... ... ... ... 9 SITE_B 82.1 0
The dimensionality of this dataframe must be: N_samples x N_Covariates
The order of samples must be identical in the covariate_matrix and the data_matrix.
After preparing both inputs, you can call harmonizationLearn to harmonize
the provided dataset.
Example usage:
>>> from neuroHarmonize import harmonizationLearn >>> import pandas as pd >>> import numpy as np >>> # load your data and all numeric covariates >>> my_data = pd.read_csv('brain_volumes.csv') >>> my_data = np.array(my_data) >>> covars = pd.read_csv('subject_info.csv') >>> # run harmonization and store the adjusted data >>> my_model, my_data_adj = harmonizationLearn(my_data, covars)
If you wish to skip the empirical Bayes step of ComBat, simply pass the optional
argument eb=False to harmonizationLearn.
If you have previously trained a harmonization model with harmonizationLearn,
you may apply the model parameters to new data with harmonizationApply.
First load the model:
>>> from neuroHarmonize import harmonizationApply, loadHarmonizationModel >>> import pandas as pd >>> import numpy as np >>> # load a pre-trained model >>> my_model = loadHarmonizationModel('../models/my_model')
Next, prepare the holdout data on which you will apply the model. This data
must look exactly like the training data for harmonizationLearn, including
the same number and order of covariates. If the holdout data contains a
different number of sites, an error will be thrown.
After preparing the holdout data simply apply the model:
>>> df_holdout = pd.read_csv('../data/brain_volumes_holdout.csv') >>> my_holdout_data = np.array(df_holdout) >>> covars = pd.read_csv('subject_info_holdout.csv') >>> my_holdout_data_adj = harmonizationApply(my_holdout_data, covars, my_model)
Note the default behavior is to run the empirical Bayes (EB) step of ComBat, which is useful for harmonizing multiple features that are similar such as genes or brain regional volumes.
To run without EB, specify eb=False in harmonizationLearn. This is
convenient when harmonizing a small number of features, e.g. fewer than 10.
You may specify nonlinear covariate effects with the optional argument:
smooth_terms. For example, you may want to specify age as a nonlinear
term in the harmonization model. This can be done easily with
harmonizationLearn:
>>> from neuroHarmonize import harmonizationLearn >>> import pandas as pd >>> import numpy as np >>> # load your data and all numeric covariates >>> my_data = pd.read_csv('brain_volumes.csv') >>> my_data = np.array(my_data) >>> covars = pd.read_csv('subject_info.csv') >>> # run harmonization with NONLINEAR effects of age >>> my_model, my_data_adj = harmonizationLearn(data, covars, smooth_terms=['AGE'])
When applying nonlinear models to holdout data, you may get an error: "some data
points fall outside the outermost knots, and I'm not sure how to handle them".
This is documented: statsmodels/statsmodels#2361.
The current workaround is to use the optional argument: smooth_term_bounds,
which controls the boundary knots for nonlinear estimation. You should specify
boundaries that contain the limits of the entire dataset, including holdout data.
This feature is currently in development.
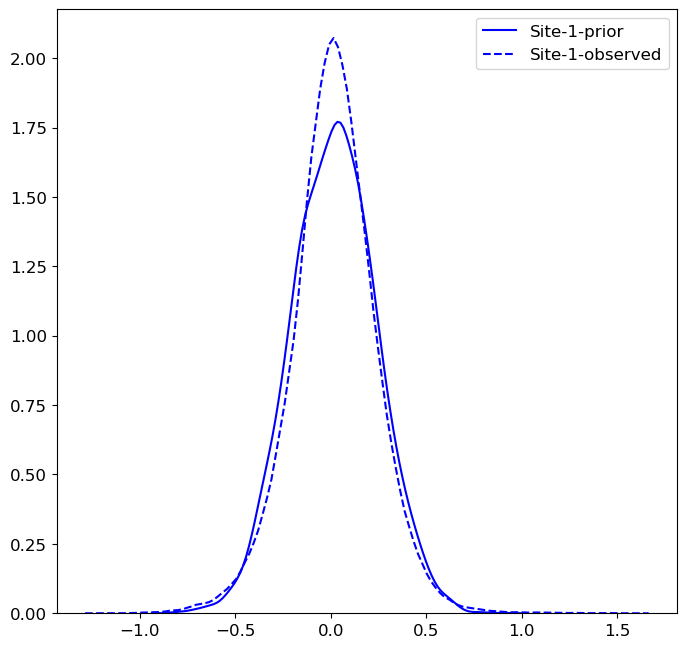
When eb=True, ComBat uses Empirical Bayes to fit a prior distribution for
the site effects for each site. You may wish to visualize fit of the prior
distribution, along with the observed distribution of site effects. The following
code example plots both distributions for the location effect of site 1.
>>> import matplotlib.pyplot as plt >>> import seaborn as sns >>> from neuroHarmonize import loadHarmonizationModel >>> model = loadHarmonizationModel('../models/my_model') >>> site_01 = stats.norm.rvs(size=10000, loc=model['gamma_bar'][0], scale=np.sqrt(model['t2'][0])) >>> sns.kdeplot(site_01, color='blue', label='Site-1-prior') >>> sns.kdeplot(model['gamma_hat'][0, :], color='blue', label='Site-1-observed', linestyle='--') >>> plt.show()
| [1] | Pomponio, R., Shou, H., Davatzikos, C., et al., (2019). "Harmonization of large MRI datasets for the analysis of brain imaging patterns throughout the lifespan." Neuroimage 208. https://doi.org/10.1016/j.neuroimage.2019.116450. |
| [2] | Fortin, J. P., N. Cullen, Y. I. Sheline, W. D. Taylor, I. Aselcioglu, P. A. Cook, P. Adams, C. Cooper, M. Fava, P. J. McGrath, M. McInnis, M. L. Phillips, M. H. Trivedi, M. M. Weissman and R. T. Shinohara (2017). "Harmonization of cortical thickness measurements across scanners and sites." Neuroimage 167: 104-120. https://doi.org/10.1016/j.neuroimage.2017.11.024. |
| [3] | W. Evan Johnson and Cheng Li, Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics, 8(1):118-127, 2007. https://doi.org/10.1093/biostatistics/kxj037. |