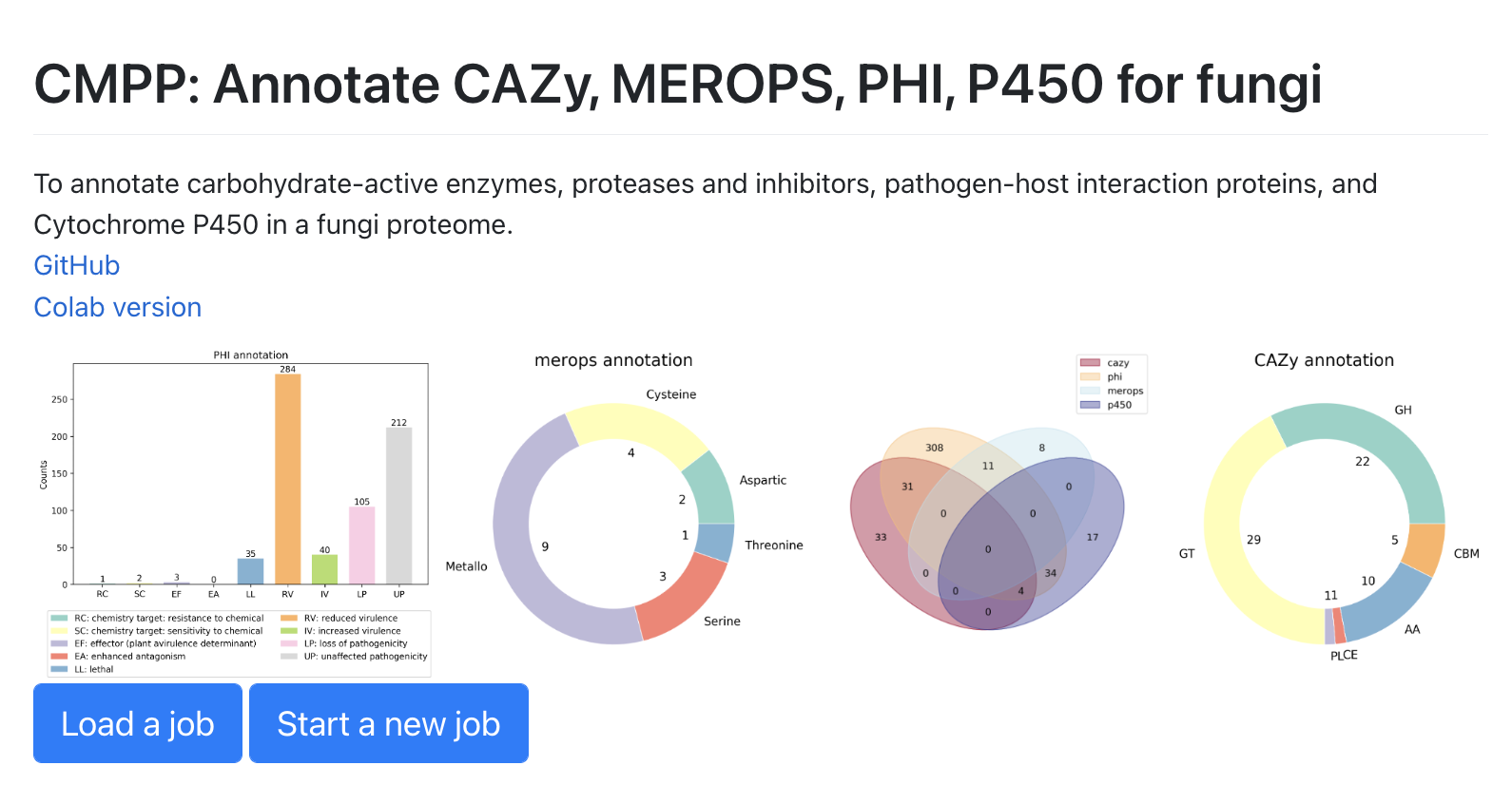
Annotate C(CAZy)M(MEROPS)P(PHI)P(P450) for fungi.
Dependency: ncbi-blast (v2.9 or higher), hmmscan, venn and functional
blastp need to support v5 database, hmmscan is part of HMMER and install venn using pip.
Clone repo to local and download functional database.
git clone https://github.com/JinyuanSun/CMPP.git
cd CMPP
wget http://185.201.226.155/CMPP/functional.tar.gz
tar vxzf functional.tar.gz./run_CMPP_anno.sh -i $protein.fas -d functionalCheck the output in CMPP_out.
A web app for biologists link
Simple cilck the start a new job button and upload a fasta file.
It also available via CoLab.
In this CoLab notebook, you can upload a custom fasta file conatining a proteome or directly download a proteome from NCBI.
- Add Signal peptide and TMHMM prediction for proteases and CAZy.