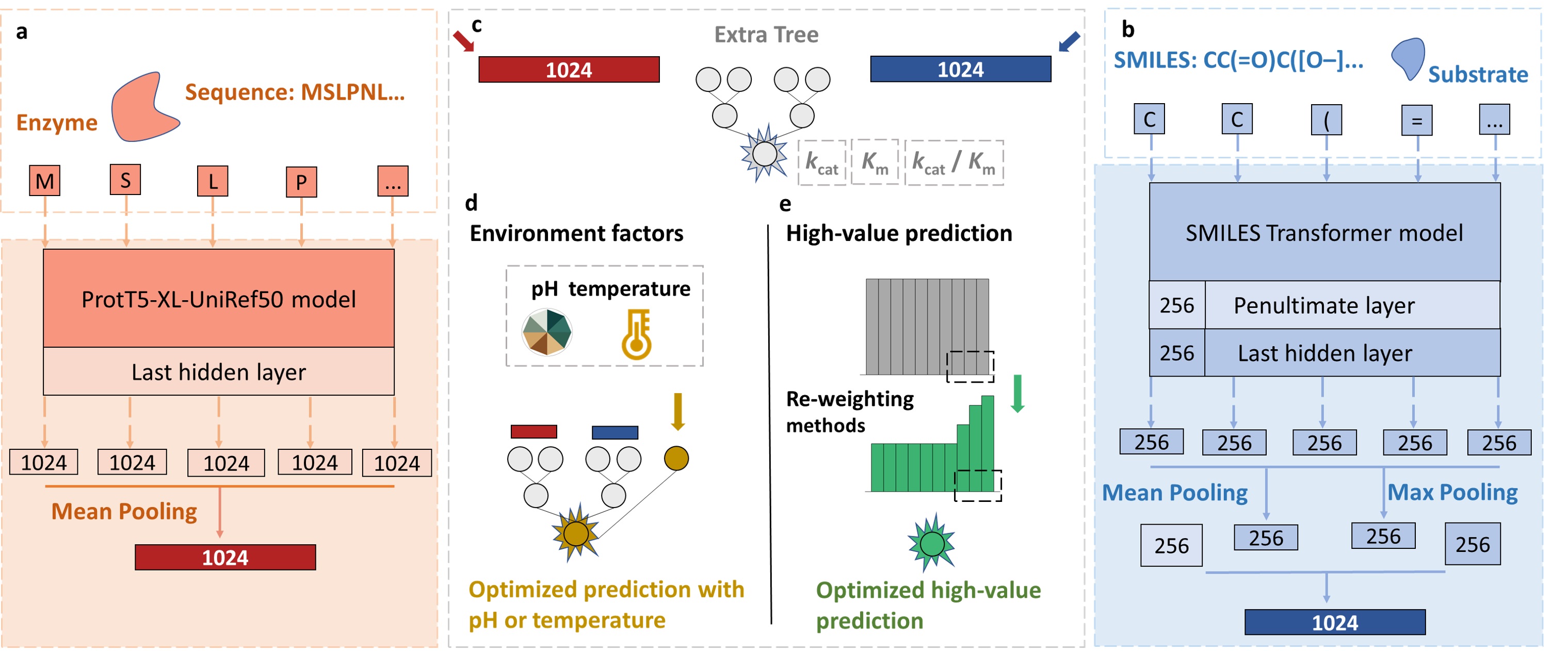
Introduction of UniKP.
Accurate enzyme kinetic parameters prediction enables optimization of reaction conditions, increasing catalytic efficiency, yield and specificity, and improving overall process economics in enzyme-dependent industries such as pharmaceuticals, food, and chemicals.The lack of a unified framework and reliable prediction tools capable of handling diverse tasks limits their practical application in numerous fields, where accurate predictions of enzyme properties can significantly impact research outcomes.Here, we introduced UniKP, a unified framework based on pretrained language models for highly accurate enzyme kcat, Km and kcat / Km prediction from protein sequences and substrate structures.
Here is the framework of UniKP.
-
For users who want to know what to expect in this project, as follows:
- (1). Out the kcat values given protein sequences and substrate structures.
- (2). Out the Km values given protein sequences and substrate structures.
- (3). Out the kcat / Km values given protein sequences and substrate structures.
| Input_v1 | Input_v2 | Model | Output |
|---|---|---|---|
| MSELMKLSAV...MAQR | CC(O)O | UniKP for kcat | 2.75 s-1 |
| MSELMKLSAV...MAQR | CC(O)O | UniKP for Km | 0.36 mM |
| MSELMKLSAV...MAQR | CC(O)O | UniKP for kcat / Km | 9.51 s-1 * mM-1 |
Notice:
- You need install pretrained protein language modoel ProtT5-XL-UniRef50 to generate enzyme representation, the link is provided on ProtT5-XL-U50.
- You need install pretrained molecular language modoel SMILES Transformer to generate substrate representation, the link is provided on SMILES Transformer.
- You also need install model UniKP for kcat, Km and kcat / Km to predict corresponding kinetic parameters, the link is provided on UniKP_model.
other packages:
- Python v3.6.9 (Anaconda installation recommended)
- PyTorch v1.10.1+cu113
- pandas v1.1.5
- NumPy v1.19.5
-
For users who want to use the deep learning model for prediction, please run these command lines at the terminal:
-
(1). Download the UniKP package
git clone https://github.com/Luo-SynBioLab/UniKP -
(2). Download required Python package
pip install numpy pandas PyTorch v1.10.1+cu113
-
-
Example for how to predict enzyme kinetic parameters from enzyme sequences and substrate structures by language model, UniKP:
import torch
from build_vocab import WordVocab
from pretrain_trfm import TrfmSeq2seq
from utils import split
from transformers import T5EncoderModel, T5Tokenizer
import re
import gc
import numpy as np
import pandas as pd
import pickle
import math
def smiles_to_vec(Smiles):
pad_index = 0
unk_index = 1
eos_index = 2
sos_index = 3
mask_index = 4
vocab = WordVocab.load_vocab('UniKP/vocab.pkl')
def get_inputs(sm):
seq_len = 220
sm = sm.split()
if len(sm)>218:
print('SMILES is too long ({:d})'.format(len(sm)))
sm = sm[:109]+sm[-109:]
ids = [vocab.stoi.get(token, unk_index) for token in sm]
ids = [sos_index] + ids + [eos_index]
seg = [1]*len(ids)
padding = [pad_index]*(seq_len - len(ids))
ids.extend(padding), seg.extend(padding)
return ids, seg
def get_array(smiles):
x_id, x_seg = [], []
for sm in smiles:
a,b = get_inputs(sm)
x_id.append(a)
x_seg.append(b)
return torch.tensor(x_id), torch.tensor(x_seg)
trfm = TrfmSeq2seq(len(vocab), 256, len(vocab), 4)
trfm.load_state_dict(torch.load('UniKP/trfm_12_23000.pkl'))
trfm.eval()
x_split = [split(sm) for sm in Smiles]
xid, xseg = get_array(x_split)
X = trfm.encode(torch.t(xid))
return X
def Seq_to_vec(Sequence):
for i in range(len(Sequence)):
if len(Sequence[i]) > 1000:
Sequence[i] = Sequence[i][:500] + Sequence[i][-500:]
sequences_Example = []
for i in range(len(Sequence)):
zj = ''
for j in range(len(Sequence[i]) - 1):
zj += Sequence[i][j] + ' '
zj += Sequence[i][-1]
sequences_Example.append(zj)
tokenizer = T5Tokenizer.from_pretrained("UniKP/prot_t5_xl_uniref50", do_lower_case=False)
model = T5EncoderModel.from_pretrained("UniKP/prot_t5_xl_uniref50")
gc.collect()
print(torch.cuda.is_available())
# 'cuda:0' if torch.cuda.is_available() else
device = torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')
model = model.to(device)
model = model.eval()
features = []
for i in range(len(sequences_Example)):
print('For sequence ', str(i+1))
sequences_Example_i = sequences_Example[i]
sequences_Example_i = [re.sub(r"[UZOB]", "X", sequences_Example_i)]
ids = tokenizer.batch_encode_plus(sequences_Example_i, add_special_tokens=True, padding=True)
input_ids = torch.tensor(ids['input_ids']).to(device)
attention_mask = torch.tensor(ids['attention_mask']).to(device)
with torch.no_grad():
embedding = model(input_ids=input_ids, attention_mask=attention_mask)
embedding = embedding.last_hidden_state.cpu().numpy()
for seq_num in range(len(embedding)):
seq_len = (attention_mask[seq_num] == 1).sum()
seq_emd = embedding[seq_num][:seq_len - 1]
features.append(seq_emd)
features_normalize = np.zeros([len(features), len(features[0][0])], dtype=float)
for i in range(len(features)):
for k in range(len(features[0][0])):
for j in range(len(features[i])):
features_normalize[i][k] += features[i][j][k]
features_normalize[i][k] /= len(features[i])
return features_normalize
if __name__ == '__main__':
sequences = ['MEDIPDTSRPPLKYVKGIPLIKYFAEALESLQDFQAQPDDLLISTYPKSGTTWVSEILDMIYQDGDVEKCRRAPVFIRVPFLEFKA'
'PGIPTGLEVLKDTPAPRLIKTHLPLALLPQTLLDQKVKVVYVARNAKDVAVSYYHFYRMAKVHPDPDTWDSFLEKFMAGEVSYGSW'
'YQHVQEWWELSHTHPVLYLFYEDMKENPKREIQKILKFVGRSLPEETVDLIVQHTSFKEMKNNSMANYTTLSPDIMDHSISAFMRK'
'GISGDWKTTFTVAQNERFDADYAKKMEGCGLSFRTQL']
Smiles = ['OC1=CC=C(C[C@@H](C(O)=O)N)C=C1']
seq_vec = Seq_to_vec(sequences)
smiles_vec = smiles_to_vec(Smiles)
fused_vector = np.concatenate((smiles_vec, seq_vec), axis=1)
# For kcat
with open('UniKP/UniKP for kcat.pkl', "rb") as f:
model = pickle.load(f)
# For Km
# with open('UniKP/UniKP for Km.pkl', "rb") as f:
# model = pickle.load(f)
# For kcat/Km
# with open('UniKP/UniKP for kcat_Km.pkl', "rb") as f:
# model = pickle.load(f)
Pre_label = model.predict(fused_vector)
Pre_label_pow = [math.pow(10, Pre_label[i]) for i in range(len(Pre_label))]
print(len(Pre_label))
res = pd.DataFrame({'sequences': sequences, 'Smiles': Smiles, 'Pre_label': Pre_label})
res.to_excel('Kinetic_parameters_predicted_label.xlsx')
- Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen 518055, China:
| Han Yu | Huaxiang Deng | Jiahui He | Jay D. Keasling | Xiaozhou Luo |
|---|
We would like to acknowledge the support from National Key R&D Program of China (2018YFA0903200), National Natural Science Foundation of China (32071421), Guangdong Basic and Applied Basic Research Foundation (2021B1515020049), Shenzhen Science and Technology Program (ZDSYS20210623091810032 and JCYJ20220531100207017), and Shenzhen Institute of Synthetic Biology Scientific Research Program (ZTXM20203001).
GNU General Public License version 3
If you use this code or our models for your publication, please cite the original paper:
Han Yu, Huaxiang Deng, Jiahui He et al. Highly accurate enzyme turnover number prediction and enzyme engineering with PreKcat, 18 May 2023, PREPRINT (Version 1) available at Research Square [https://doi.org/10.21203/rs.3.rs-2749688/v1]