check here for a chinese tutorial.
Calculate the RMSD between two antibody structure (including nanobody and antibody).
First, install abnumber using conda.
conda install -c bioconda abnumberThen, install ab_rmsd from github.
pip install git+https://github.com/pengzhangzhi/ab_rmsd.gitIf you want to calculate the DockQ between complex structures, run the following command.
cd DockQ
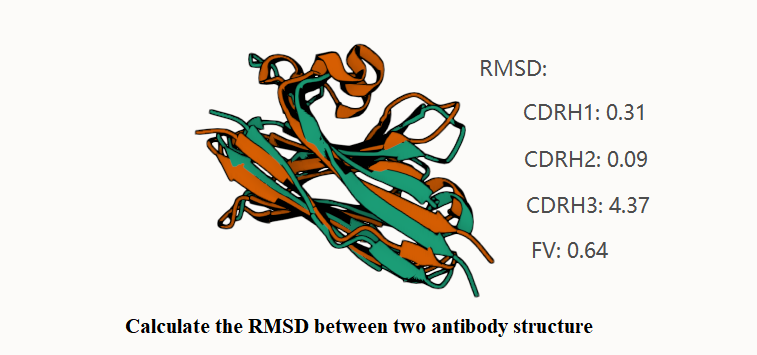
makeCalculate the RMSD between predicted and native nanobody structure.
from ab_rmsd import calc_ab_rmsd
# two example pdb files are provided in the `example` folder.
native = "example/7d6y_1_B.pdb"
pred = "example/pred_7d6y_1_B.pdb"
rmsd = calc_ab_rmsd(native,pred)
print(rmsd)
"""
output:
{'CDRH1': tensor(0.3136), 'CDRH2': tensor(0.0898), 'CDRH3': tensor(4.3704), 'fv-H': tensor(0.6426)}
"""In the terminal:
abrmsd --pred pred_7d6y_1_B.pdb --native 7d6y_1_B.pdb --verbose
# Output:
[INFO] Renumbered chain B (H)
[INFO] Renumbered chain A (H)
_ _ ____ __ __ ____ ____
/ \ | |__ | _ \| \/ / ___|| _ \
/ _ \ | '_ \ | |_) | |\/| \___ \| | | |
/ ___ \| |_) | | _ <| | | |___) | |_| |
/_/ \_\_.__/ |_| \_\_| |_|____/|____/
>>> Result
Frag RMSD(Å)
CDRH1 0.3136
CDRH2 0.0898
CDRH3 4.3704
fv-H 0.6426
>>> EndCalculate the RMSD between paired antibody structures (containing heavy and light chains).
from ab_rmsd import calc_ab_rmsd
# two example pdb files are provided in the `exampl`e folder.
native = "example/7s0b_.pdb"
pred = "example/pred_7s0b_.pdb"
rmsd = calc_ab_rmsd(native,pred)
print(rmsd)
"""
output:
{
'CDRH1': tensor(25.6173), 'CDRH2': tensor(15.5819), 'CDRH3': tensor(25.7562), 'fv-H': tensor(15.9964),
'CDRL1': tensor(11.8419), 'CDRL2': tensor(13.8057), 'CDRL3': tensor(17.1446), 'fv-L': tensor(15.9478)
}
"""Calculate the DockQ scores between predicted and native complex structure.
from DockQ.DockQ import calc_DockQ
scores = calc_DockQ(model='example/pred_7s0b_.pdb',native='example/7s0b_.pdb')
print(scores)
"""
The first four scores are usually used to evaluate the docking performance.
{
'DockQ': 0.011549873197136384,
'irms': 17.429353912635577,
'Lrms': 50.73969606449461,
'fnat': 0.0,
'nat_correct': 0, 'nat_total': 55, 'fnonnat': 1.0,
'nonnat_count': 9, 'model_total': 9,
'chain1': 'A', 'chain2': 'B', 'len1': 121,
'len2': 107, 'class1': 'receptor', 'class2': 'ligand'
}
"""Use the cli to calculate the DockQ score.
./DockQ/DockQ.py example/pred_7s0b_.pdb example/7s0b_.pdbaddDockQas an evaluation for heavy and light chain complex structure.for multi-chain antibody, superimpose each chain seperately and calculate the RMSD. instead of superimposing the complex structure.
- Part of the code is adapted from shitong's Diffab.
- Code about the DockQ is from https://github.com/bjornwallner/DockQ.
- Huge thanks to @JinyuanSun for contributing the cli tool.