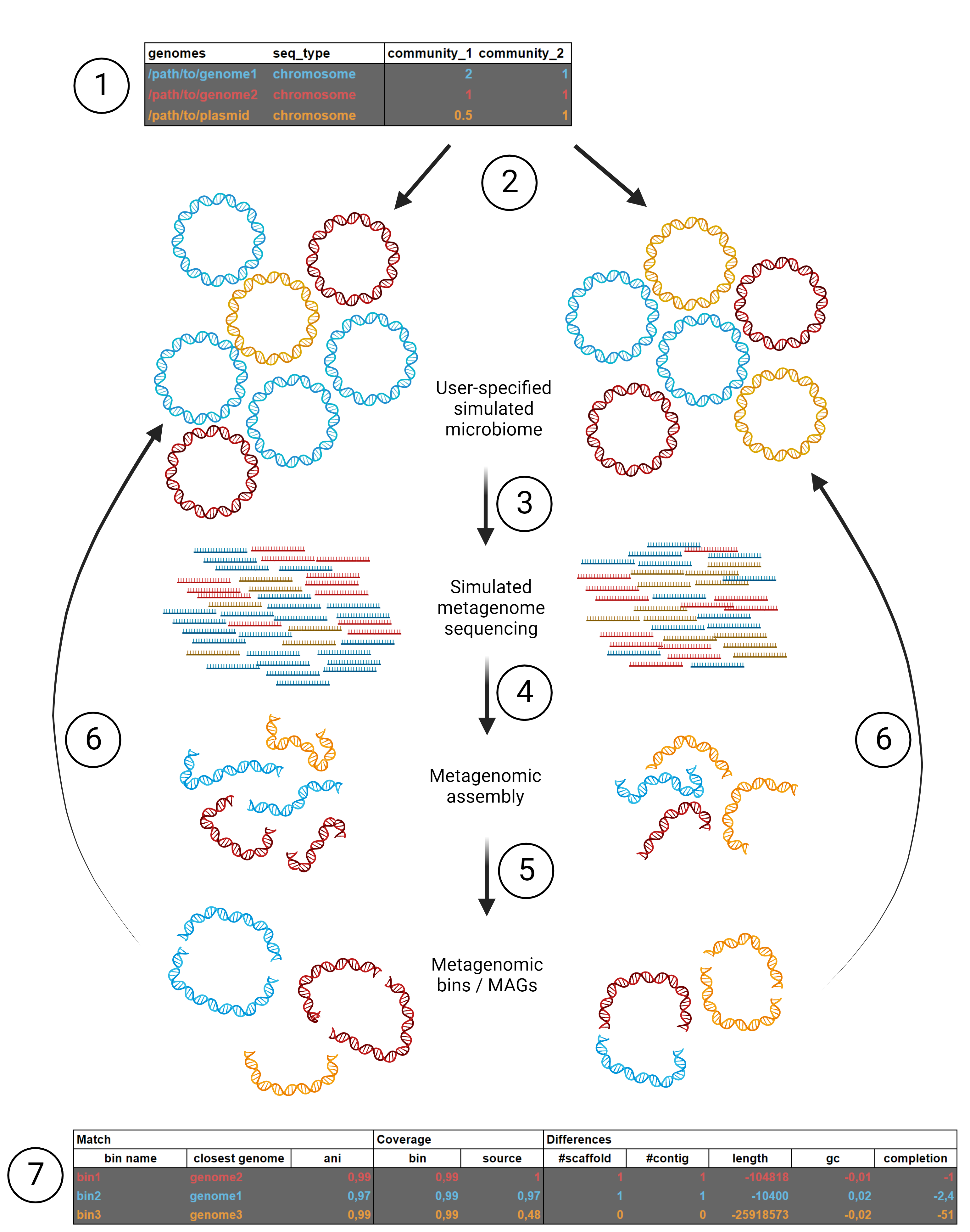
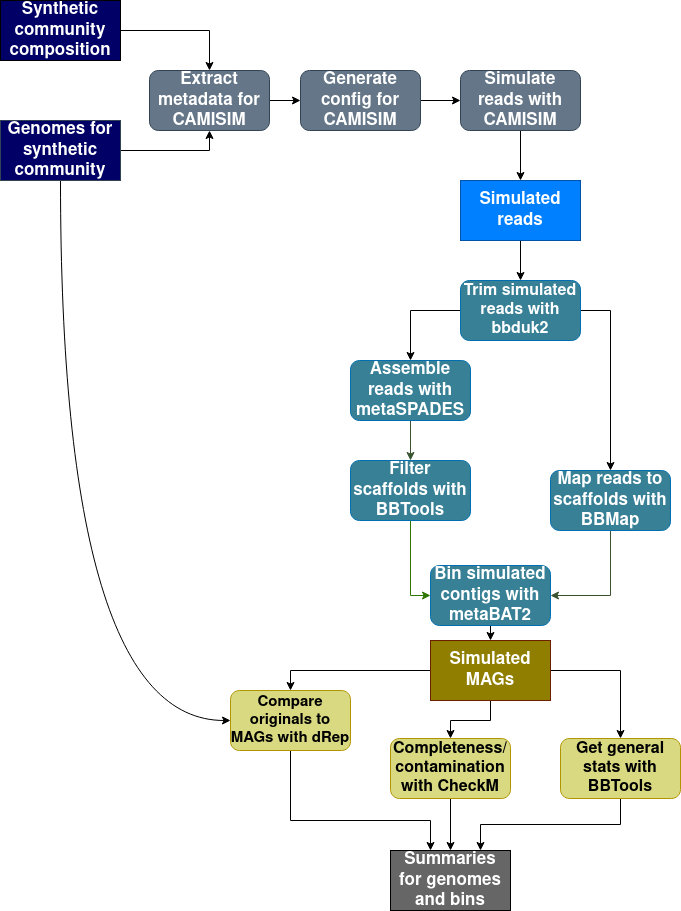
MAGICIAN is a tool for easily generating simulated metagenome-assembled genomes from a user-determined "community".
MAGICIAN is a Snakemake pipeline that uses conda or mamba to manage dependencies.
Thus, it primarily requires Snakemake and conda or mamba to be used (mamba is recommended).
Requirements for the base environment are given in requirements.yml and can be installed as follows:
Using conda:
conda env create --file requirements.yml
Using mamba:
mamba env create --file requirements.yml
It is also necessary to install a fork of CAMISIM 1.2 in order to use custom error profiles. This can be done by running
git clone https://github.com/KatSteinke/CAMISIM
In order to get started with MAGICIAN, simply clone the repository:
git clone https://github.com/KatSteinke/magician
You will also have to adapt the config file given under config/default_config.yml.
Change the path given under camisim_path in default_config.ymlto the path to your forked copy of CAMISIM.
If you use mamba (recommended due to speed), change the setting for conda_frontend to mamba.
MAGICIAN requires the following files to run:
- genome sequences of the organisms the simulated community should consist of, in genbank or fasta format
- a tab-separated file of sample distributions. The first column lists the paths to the genomes, the second lists sequence type (chromosome or plasmid) and all subsequent columns list community composition names and the relative abundance of the sequences in these communities:
| genomes | seq_type | community1 | community2 | ... |
|------------------|------------|------------|------------|-----|
| /path/to/genome1 | chromosome | 1 | 1.5 | |
| /path/to/genome2 | chromosome | 1 | 0 | |
| /path/to/plasmid | plasmid | 1 | 1 | |
MAGICIAN is started using run_magician.py:
run_magician.py [-h] [--target TARGET]
[--profile_type {mbarc,hi,mi,hi150,own}]
[--profile_name PROFILE_NAME]
[--profile_readlength PROFILE_READLENGTH]
[--insert_size INSERT_SIZE] [--cluster CLUSTER]
[--config_file CONFIG_FILE]
[--cores CORES]
community_file
[--snake_flags "SNAKE_FLAGS..."]
community_file: the tab-separated file with sample distributions for the community/communities you wish to simulate.--snake_flags: the flags to be passed on to Snakemake, enclosed in double quotes. For a dry run, use"-n ". For all else, refer to Snakemake's documentation.
-
--target: the desired output file or rule. By default, MAGICIAN runs the entire workflow for all communities in the input file given. To run the workflow for a single community, givesummaries/bin_summary_[COMMUNITY].xlsxhere, replacing[COMMUNITY]with the name of the community you wish to simulate. -
--profile_type: the error profile CAMISIM should use for ART. This defaults to CAMISIM's default ofmbarc; other choices arehi,mi,hi150,own. The last allows users to specify their own profiles. -
--profile_name: required when specifying one's own profile. This is the base path to the forward/reverse reads' error profiles (e.g.path/to/custom/profile_Rif forward and reverse reads are located atpath/to/custom/profile_R1.txtandpath/to/custom/profile_R2.txtrespectively) -
--profile_readlength: the read length used for the custom error profile; required when specifying one's own error profile. -
--insert_size: mean insert size for read simulation (defaults to 270 bp) -
--cluster: when using Snakemake's cluster mode, supply the command for submitting jobs as you would with Snakemake -
--cores: the amount of cores Snakemake should use (default: 6) -
--config_file: the path to the configuration file to use, if not using the default filedefault_config.yml
To start a test run with the sample genomes found in test/data/test_genomes, run python3 run_magician.py without any arguments. The script will show usage and ask whether to start a test run:
usage: run_magician.py [-h] [--target TARGET] [--profile_type {mbarc,hi,mi,hi150,own}]
[--profile_name PROFILE_NAME]
[--profile_readlength PROFILE_READLENGTH]
[--insert_size INSERT_SIZE] [--cluster CLUSTER]
[--config_file CONFIG_FILE]
[--cores CORES]
[--snake_flags [SNAKE_FLAGS ...]]
community_file
Start local example run with sample genomes and output to /home/kma/magician? [y/n]
Remember to edit config/default_config.yml to specify your CAMISIM installation.
Confirm with y to start the test run.
Example summary files for such a run can be found under test/data/sample_summaries; the full output is available at Zenodo.
Copyright 2023 Kat Steinke
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this work except in compliance with the License. You may obtain a copy of the License at
http://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.