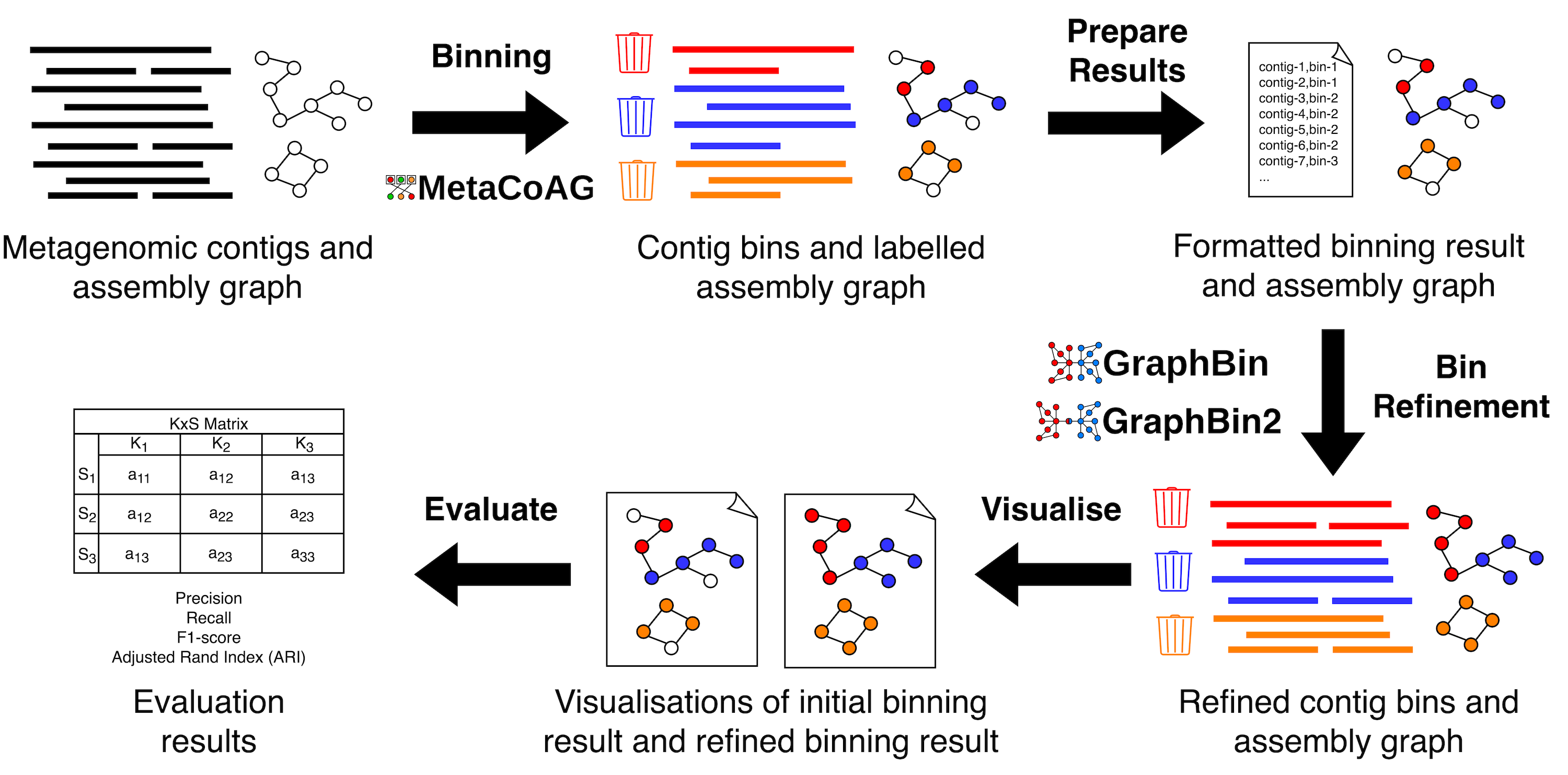
GraphBin-Tk combines assembly graph-based metagenomic bin-refinement and binning techniques GraphBin, GraphBin2 and MetaCoAG along with support functionality to visualise and evaluate results, into one comprehensive toolkit.
Detailed documentation hosted at Read the Docs is coming soon!
# clone repository
git clone https://github.com/metagentools/gbintk.git
# move to gbintk directory
cd gbintk
# create and activate conda env
conda env create -f environment.yml
conda activate gbintk
# install using flit
flit install
# test installation
gbintk --helpRun gbintk --help or gbintk -h to list the help message for GraphBin-Tk.
Usage: gbintk [OPTIONS] COMMAND [ARGS]...
gbintk (GraphBin-Tk): Assembly graph-based metagenomic binning toolkit
Options:
-v, --version Show the version and exit.
-h, --help Show this message and exit.
Commands:
graphbin GraphBin: Refined Binning of Metagenomic Contigs using...
graphbin2 GraphBin2: Refined and Overlapped Binning of Metagenomic...
metacoag MetaCoAG: Binning Metagenomic Contigs via Composition,...
prepare Format the initial binning result from an existing binning tool
visualise Visualise binning and refinement results
evaluate Evaluate the binning results given a ground truthIf you use GraphBin-Tk in your work, please cite the relevant tools.
GraphBin
Vijini Mallawaarachchi, Anuradha Wickramarachchi, Yu Lin. GraphBin: Refined binning of metagenomic contigs using assembly graphs. Bioinformatics, Volume 36, Issue 11, June 2020, Pages 3307–3313, DOI: https://doi.org/10.1093/bioinformatics/btaa180
GraphBin2
Vijini G. Mallawaarachchi, Anuradha S. Wickramarachchi, and Yu Lin. GraphBin2: Refined and Overlapped Binning of Metagenomic Contigs Using Assembly Graphs. In 20th International Workshop on Algorithms in Bioinformatics (WABI 2020). Leibniz International Proceedings in Informatics (LIPIcs), Volume 172, pp. 8:1-8:21, Schloss Dagstuhl – Leibniz-Zentrum für Informatik (2020). DOI: https://doi.org/10.4230/LIPIcs.WABI.2020.8
Mallawaarachchi, V.G., Wickramarachchi, A.S. & Lin, Y. Improving metagenomic binning results with overlapped bins using assembly graphs. Algorithms Mol Biol 16, 3 (2021). DOI: https://doi.org/10.1186/s13015-021-00185-6
MetaCoAG
Mallawaarachchi, V., Lin, Y. (2022). MetaCoAG: Binning Metagenomic Contigs via Composition, Coverage and Assembly Graphs. In: Pe'er, I. (eds) Research in Computational Molecular Biology. RECOMB 2022. Lecture Notes in Computer Science(), vol 13278. Springer, Cham. DOI: https://doi.org/10.1007/978-3-031-04749-7_5
Vijini Mallawaarachchi and Yu Lin. Accurate Binning of Metagenomic Contigs Using Composition, Coverage, and Assembly Graphs. Journal of Computational Biology 2022 29:12, 1357-1376. DOI: https://doi.org/10.1089/cmb.2022.0262
GraphBin-Tk is funded by an Essential Open Source Software for Science Grant from the Chan Zuckerberg Initiative.