The trendbreaker package implements tools for detecting changes in temporal trends of a single response variable. It provides a coherent interface to several modeling tools, alongside functions for model selection and outlier detection. It implements the Automatic Selection of Models and Outlier Detection for Epidemmics (ASMODEE), an algorithm originally designed for detecting changes in COVID-19 case incidence.
ASMODEE proceeds by:
- defining a training set excluding the last k data points
- identifying the temporal trend in the training set by fitting a range of (user-specified) models to the data and retaining the best predicting / fitting model
- calculating the prediction interval (PI) of the temporal trend
- classifying any data point outside the PI as outlier
The algorithm can be applied with fixed, user-specified value of k, so as to monitor potential changes in this recent time period. Alternatively, the optimal value of k can be determined automatically.
Disclaimer: this is work in progress. Please reach out to the authors before using this package. Also note this package may soon be renamed to avoid clashes with other projects and reflect a more general scope.
Once it is released on CRAN, you will be able to install the stable version of the package with:
install.packages("trendbreaker")The development version can be installed from GitHub with:
if (!require(remotes)) {
install.packages("remotes")
}
remotes::install_github("reconhub/trendbreaker")The best place to start for using this package is to read the
documentation of the function asmodee and run its example:
library(trendbreaker)
?asmodee
example(asmodee)The package implements the following main functions
-
asmodee: implements the Automatic Selection of Models and Outlier DEtection for Epidemics -
select_model: a function to select the best-fitting/best-predicting model from a range of user-specified models -
detect_changepoint: a function to detect the points at which recent data deviate from previous temporal trends using a fitted model and data -
detect_outliers: a function to identify outliers using a fitted model and data
We illustrate ASMODEE using publicly available NHS pathways data
recording self-reporting of potential COVID-19 cases in England (see
?nhs_pathways_covid19 for more information). ASMODEE would typically
be more useful to investigate shifts in temporal trends from a large
number of time series (e.g. at a fine geographic scale), but is this is
purely for illustrative purposes, we use the full dataset for England.
See ?asmodee for further example stratified by geographic units.
library(trendbreaker)
library(tidyverse)
#> ── Attaching packages ─────────────────────────────────────── tidyverse 1.3.0 ──
#> ✔ ggplot2 3.3.1 ✔ purrr 0.3.4
#> ✔ tibble 3.0.1 ✔ dplyr 1.0.0
#> ✔ tidyr 1.1.0 ✔ stringr 1.4.0
#> ✔ readr 1.3.1 ✔ forcats 0.5.0
#> ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
# load data
data(nhs_pathways_covid19)
# select last 6 weeks of data
first_date <- max(nhs_pathways_covid19$date, na.rm = TRUE) - 6*7
pathways_recent <- nhs_pathways_covid19 %>%
filter(date >= first_date)
# define candidate models
models <- list(
regression = lm_model(count ~ day),
poisson_constant = glm_model(count ~ 1, family = "poisson"),
negbin_time = glm_nb_model(count ~ day),
negbin_time_weekday = glm_nb_model(count ~ day + weekday)
)
# analyses on all data
counts_overall <- pathways_recent %>%
group_by(date, day, weekday) %>%
summarise(count = sum(count))
#> `summarise()` regrouping output by 'date', 'day' (override with `.groups` argument)
# results with automated detection of 'k'
res <- asmodee(counts_overall, models, method = evaluate_aic)
res
#> $k
#> [1] 7
#>
#> $model
#> $model
#>
#> Call: MASS::glm.nb(formula = count ~ day + weekday, data = data, init.theta = 76.80468966,
#> link = log)
#>
#> Coefficients:
#> (Intercept) day weekdaymonday weekdayweekend
#> 10.81062 -0.02057 0.24398 -0.11588
#>
#> Degrees of Freedom: 35 Total (i.e. Null); 32 Residual
#> Null Deviance: 186.5
#> Residual Deviance: 36.09 AIC: 665.4
#>
#> $predict
#> function (newdata, alpha = 0.05)
#> {
#> suppressWarnings(res <- add_prediction_interval(data = newdata,
#> model = model, alpha = alpha))
#> col_name <- as.character(formula[[2]])
#> append_observed_column(res, res[[col_name]])
#> }
#> <bytecode: 0x55aca770b160>
#> <environment: 0x55acad36c0a8>
#>
#> attr(,"class")
#> [1] "trendbreaker_model_fit" "list"
#>
#> $n_outliers
#> [1] 9
#>
#> $n_outliers_train
#> [1] 2
#>
#> $n_outliers_recent
#> [1] 7
#>
#> $p_value
#> [1] 4.076898e-05
#>
#> $results
#> # A tibble: 43 x 10
#> # Groups: date, day [43]
#> date day weekday count pred lower upper observed outlier
#> <date> <int> <fct> <int> <dbl> <dbl> <dbl> <int> <lgl>
#> 1 2020-04-16 29 rest_o… 29497 27288. 21520 33729 29497 FALSE
#> 2 2020-04-17 30 rest_o… 27007 26733. 21082 33043 27007 FALSE
#> 3 2020-04-18 31 weekend 25453 23323. 18392 28829 25453 FALSE
#> 4 2020-04-19 32 weekend 23387 22848. 18018 28243 23387 FALSE
#> 5 2020-04-20 33 monday 29287 32078. 25299 39648 29287 FALSE
#> 6 2020-04-21 34 rest_o… 23134 24621. 19417 30434 23134 FALSE
#> 7 2020-04-22 35 rest_o… 21803 24120. 19021 29815 21803 FALSE
#> 8 2020-04-23 36 rest_o… 22298 23629. 18634 29208 22298 FALSE
#> 9 2020-04-24 37 rest_o… 22027 23148. 18254 28613 22027 FALSE
#> 10 2020-04-25 38 weekend 18861 20196. 15925 24965 18861 FALSE
#> # … with 33 more rows, and 1 more variable: classification <fct>
#>
#> attr(,"class")
#> [1] "trendbreaker" "list"
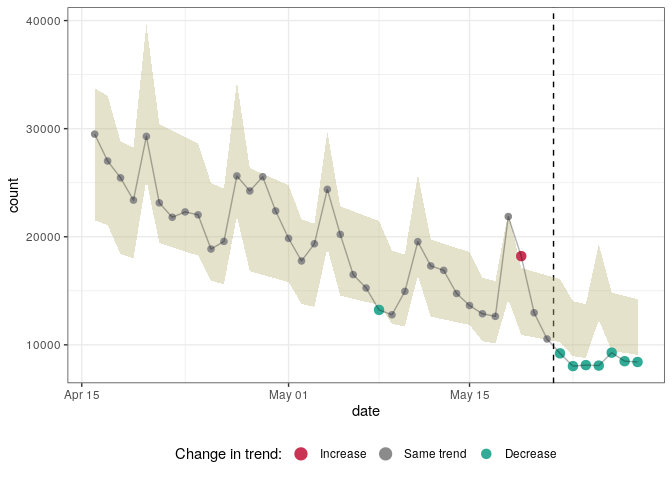
plot(res, "date")You can define a number of different regression models using a common interface. Once defined you can use different strategies to select the best-fitting/best-predicting model.
As an example we try to predict hp of the famous mtcars dataset. Of
course, this is just a toy example. Usually you would use the package to
predict counts data in a time series.
First we define some potential models:
library(trendbreaker)
stan_cache <- tempfile() # stan compile to c++ and we cache the code
models <- list(
null = lm_model(hp ~ 1),
glm_poisson = glm_model(hp ~ 1 + cyl + drat + wt + qsec + am, poisson),
lm_complex = lm_model(hp ~ 1 + cyl + drat + wt + qsec + am),
negbin_complex = glm_nb_model(hp ~ 1 + cyl + drat + wt + qsec + am),
brms_complex = brms_model(
hp ~ 1 + cyl + drat + wt + qsec + am,
family = brms::negbinomial(),
file = stan_cache
)
)Then we evaluate them using N-Fold cross validation.
# we do CV and evaluate three loss function:
# Root-mean-squared error, the huber-loss and mean absolute error.
# The package works with `yardstick` by default.
out <- capture.output( # no log output in readme :)
auto_select <- select_model(mtcars, models,
method = evaluate_resampling,
metrics = list(yardstick::rmse, yardstick::huber_loss, yardstick::mae)
)
)
auto_select$leaderboard
#> # A tibble: 5 x 4
#> model huber_loss mae rmse
#> <chr> <dbl> <dbl> <dbl>
#> 1 brms_complex 18.2 18.7 18.7
#> 2 glm_poisson 21.2 21.7 21.7
#> 3 negbin_complex 22.8 23.3 23.3
#> 4 lm_complex 26.2 26.7 26.7
#> 5 null 57.8 58.3 58.3